Gian Franco Piredda
Validation and Generalizability of Self-Supervised Image Reconstruction Methods for Undersampled MRI
Jan 29, 2022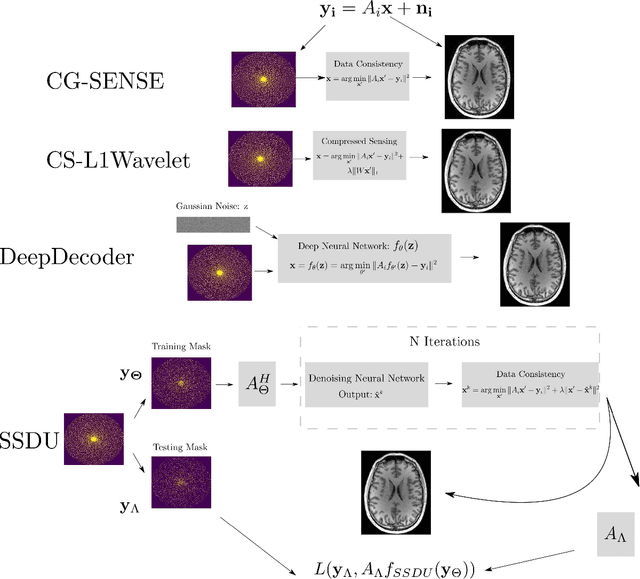
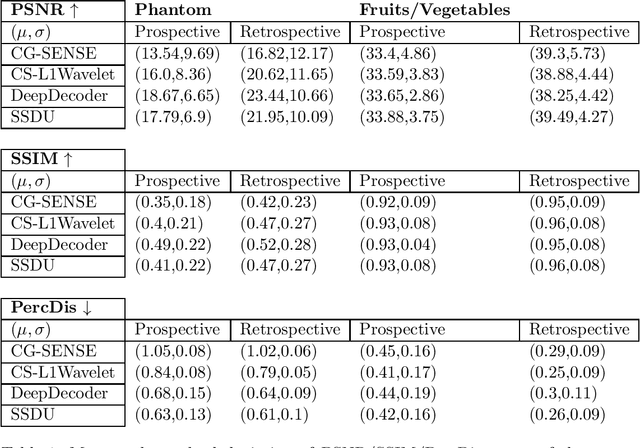
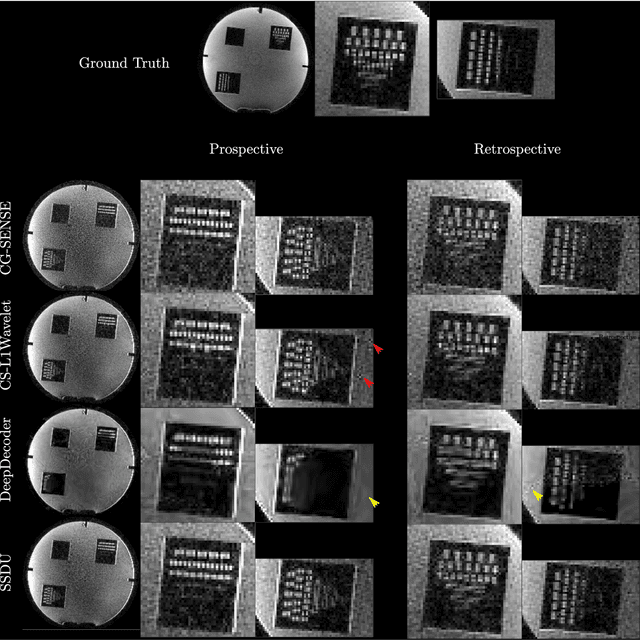
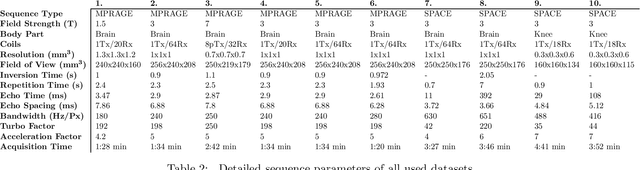
Abstract:Purpose: To investigate aspects of the validation of self-supervised algorithms for reconstruction of undersampled MR images: quantitative evaluation of prospective reconstructions, potential differences between prospective and retrospective reconstructions, suitability of commonly used quantitative metrics, and generalizability. Theory and Methods: Two self-supervised algorithms based on self-supervised denoising and neural network image priors were investigated. These methods are compared to a least squares fitting and a compressed sensing reconstruction using in-vivo and phantom data. Their generalizability was tested with prospectively under-sampled data from experimental conditions different to the training. Results: Prospective reconstructions can exhibit significant distortion relative to retrospective reconstructions/ground truth. Pixel-wise quantitative metrics may not capture differences in perceptual quality accurately, in contrast to a perceptual metric. All methods showed potential for generalization; generalizability is more affected by changes in anatomy/contrast than other changes. No-reference image metrics correspond well with human rating of image quality for studying generalizability. Compressed Sensing and learned denoising perform similarly well on all data. Conclusion: Self-supervised methods show promising results for accelerating image reconstruction in clinical routines. Nonetheless, more work is required to investigate standardized methods to validate reconstruction algorithms for future clinical use.
Multi-compartment diffusion MRI, T2 relaxometry and myelin water imaging as neuroimaging descriptors for anomalous tissue detection
Apr 15, 2021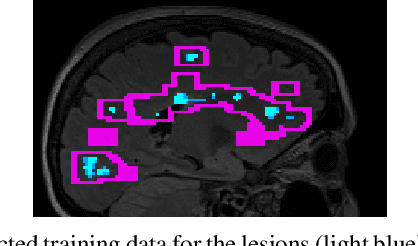
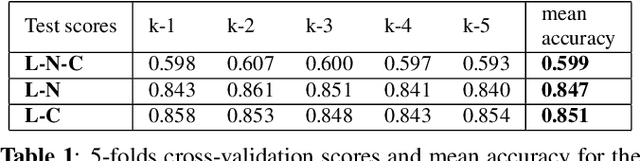
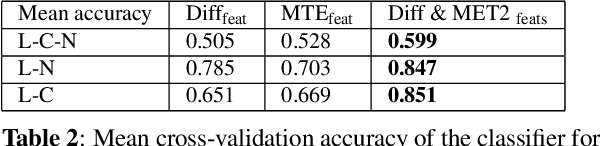
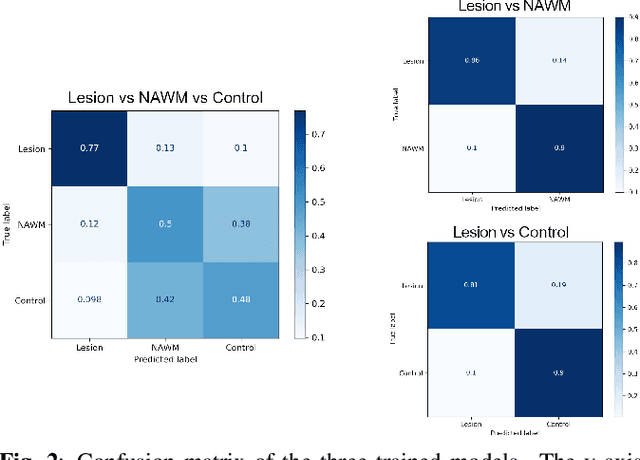
Abstract:Multiple sclerosis (MS) is an inflammatory and neurodegenerative disease characterized by diffuse and focal areas of tissue loss. Conventional MRI techniques such as T1-weighted and T2-weighted scans are generally used in the diagnosis and prognosis of the disease. Yet, these methods are limited by the lack of specificity between lesions, their perilesional area and non-lesional tissue. Alternative MRI techniques exhibit a higher level of sensitivity to focal and diffuse MS pathology than conventional MRI acquisitions. However, they still suffer from limited specificity when considered alone. In this work, we have combined tissue microstructure information derived from multicompartment diffusion MRI and T2 relaxometry models to explore the voxel-based prediction power of a machine learning model in a cohort of MS patients and healthy controls. Our results show that the combination of multi-modal features, together with a boosting enhanced decision-tree based classifier, which combines a set of weak classifiers to form a strong classifier via a voting mechanism, is able to utilise the complementary information for the classification of abnormal tissue.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge