Geoffrey Fairchild
Los Alamos National Laboratory, Los Alamos, NM, USA, The University of Iowa, Iowa City, IA, USA
"Thought I'd Share First": An Analysis of COVID-19 Conspiracy Theories and Misinformation Spread on Twitter
Dec 14, 2020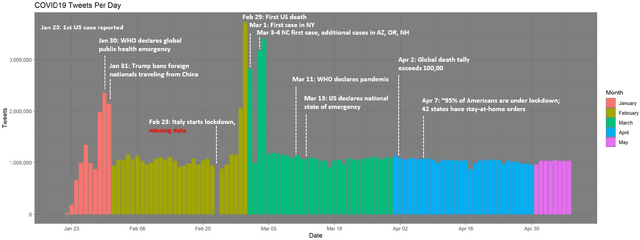
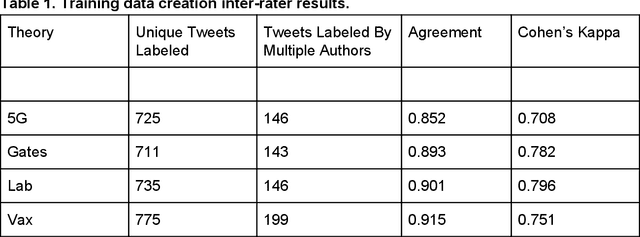
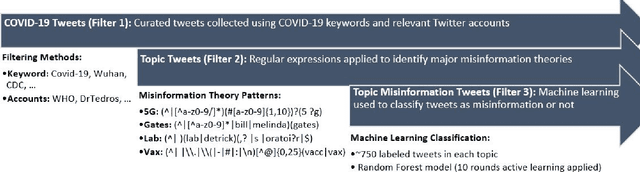
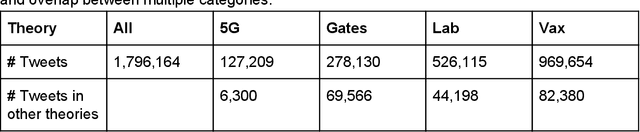
Abstract:Background: Misinformation spread through social media is a growing problem, and the emergence of COVID-19 has caused an explosion in new activity and renewed focus on the resulting threat to public health. Given this increased visibility, in-depth analysis of COVID-19 misinformation spread is critical to understanding the evolution of ideas with potential negative public health impact. Methods: Using a curated data set of COVID-19 tweets (N ~120 million tweets) spanning late January to early May 2020, we applied methods including regular expression filtering, supervised machine learning, sentiment analysis, geospatial analysis, and dynamic topic modeling to trace the spread of misinformation and to characterize novel features of COVID-19 conspiracy theories. Results: Random forest models for four major misinformation topics provided mixed results, with narrowly-defined conspiracy theories achieving F1 scores of 0.804 and 0.857, while more broad theories performed measurably worse, with scores of 0.654 and 0.347. Despite this, analysis using model-labeled data was beneficial for increasing the proportion of data matching misinformation indicators. We were able to identify distinct increases in negative sentiment, theory-specific trends in geospatial spread, and the evolution of conspiracy theory topics and subtopics over time. Conclusions: COVID-19 related conspiracy theories show that history frequently repeats itself, with the same conspiracy theories being recycled for new situations. We use a combination of supervised learning, unsupervised learning, and natural language processing techniques to look at the evolution of theories over the first four months of the COVID-19 outbreak, how these theories intertwine, and to hypothesize on more effective public health messaging to combat misinformation in online spaces.
Eliciting Disease Data from Wikipedia Articles
Aug 24, 2015

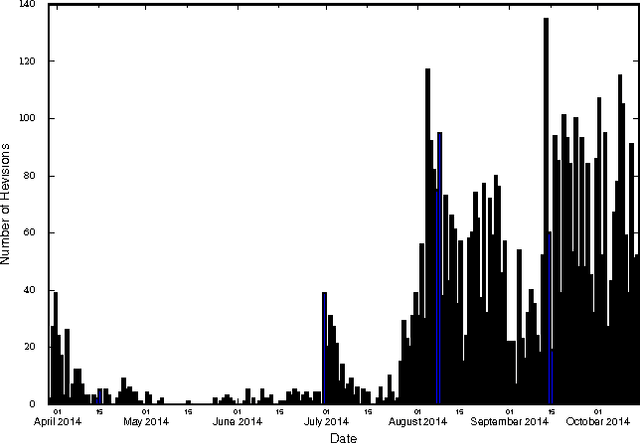

Abstract:Traditional disease surveillance systems suffer from several disadvantages, including reporting lags and antiquated technology, that have caused a movement towards internet-based disease surveillance systems. Internet systems are particularly attractive for disease outbreaks because they can provide data in near real-time and can be verified by individuals around the globe. However, most existing systems have focused on disease monitoring and do not provide a data repository for policy makers or researchers. In order to fill this gap, we analyzed Wikipedia article content. We demonstrate how a named-entity recognizer can be trained to tag case counts, death counts, and hospitalization counts in the article narrative that achieves an F1 score of 0.753. We also show, using the 2014 West African Ebola virus disease epidemic article as a case study, that there are detailed time series data that are consistently updated that closely align with ground truth data. We argue that Wikipedia can be used to create the first community-driven open-source emerging disease detection, monitoring, and repository system.
Global disease monitoring and forecasting with Wikipedia
Jul 15, 2014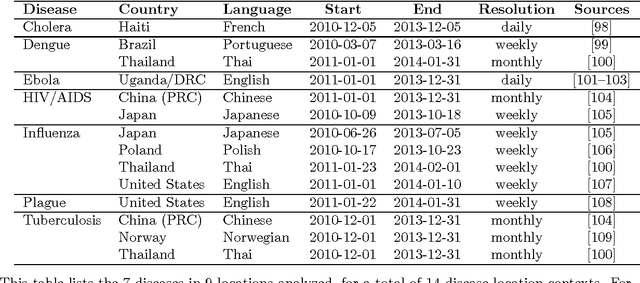
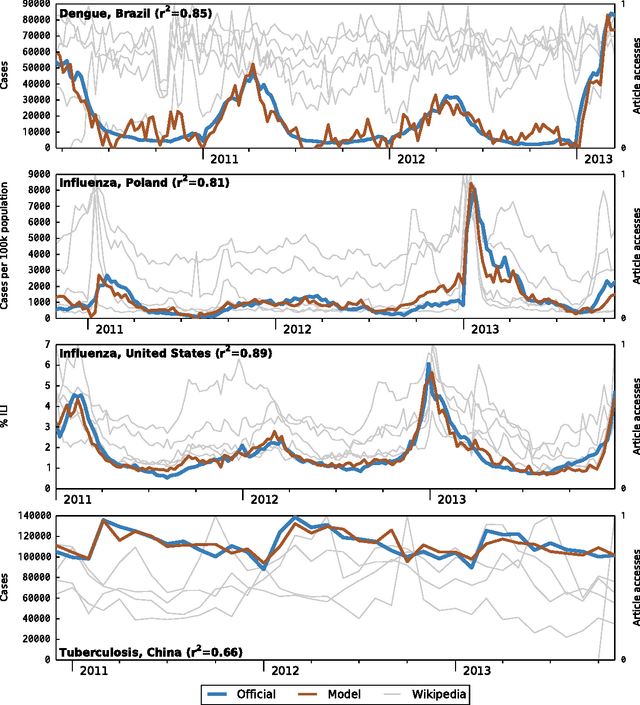

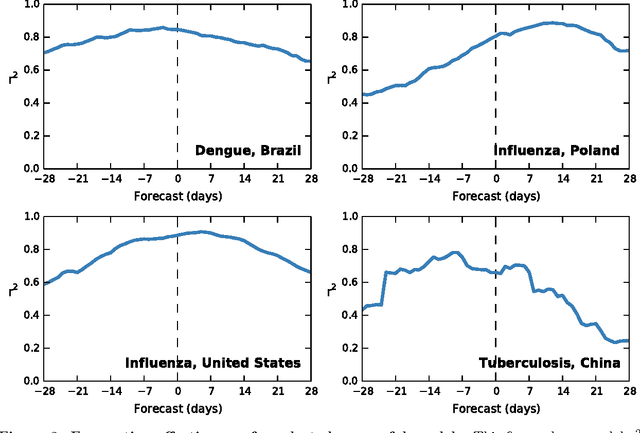
Abstract:Infectious disease is a leading threat to public health, economic stability, and other key social structures. Efforts to mitigate these impacts depend on accurate and timely monitoring to measure the risk and progress of disease. Traditional, biologically-focused monitoring techniques are accurate but costly and slow; in response, new techniques based on social internet data such as social media and search queries are emerging. These efforts are promising, but important challenges in the areas of scientific peer review, breadth of diseases and countries, and forecasting hamper their operational usefulness. We examine a freely available, open data source for this use: access logs from the online encyclopedia Wikipedia. Using linear models, language as a proxy for location, and a systematic yet simple article selection procedure, we tested 14 location-disease combinations and demonstrate that these data feasibly support an approach that overcomes these challenges. Specifically, our proof-of-concept yields models with $r^2$ up to 0.92, forecasting value up to the 28 days tested, and several pairs of models similar enough to suggest that transferring models from one location to another without re-training is feasible. Based on these preliminary results, we close with a research agenda designed to overcome these challenges and produce a disease monitoring and forecasting system that is significantly more effective, robust, and globally comprehensive than the current state of the art.
* 27 pages; 4 figures; 4 tables. Version 2: Cite McIver & Brownstein and adjust novelty claims accordingly; revise title; various revisions for clarity
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge