Gary K. Owens
ProtoGMM: Multi-prototype Gaussian-Mixture-based Domain Adaptation Model for Semantic Segmentation
Jun 27, 2024



Abstract:Domain adaptive semantic segmentation aims to generate accurate and dense predictions for an unlabeled target domain by leveraging a supervised model trained on a labeled source domain. The prevalent self-training approach involves retraining the dense discriminative classifier of $p(class|pixel feature)$ using the pseudo-labels from the target domain. While many methods focus on mitigating the issue of noisy pseudo-labels, they often overlook the underlying data distribution p(pixel feature|class) in both the source and target domains. To address this limitation, we propose the multi-prototype Gaussian-Mixture-based (ProtoGMM) model, which incorporates the GMM into contrastive losses to perform guided contrastive learning. Contrastive losses are commonly executed in the literature using memory banks, which can lead to class biases due to underrepresented classes. Furthermore, memory banks often have fixed capacities, potentially restricting the model's ability to capture diverse representations of the target/source domains. An alternative approach is to use global class prototypes (i.e. averaged features per category). However, the global prototypes are based on the unimodal distribution assumption per class, disregarding within-class variation. To address these challenges, we propose the ProtoGMM model. This novel approach involves estimating the underlying multi-prototype source distribution by utilizing the GMM on the feature space of the source samples. The components of the GMM model act as representative prototypes. To achieve increased intra-class semantic similarity, decreased inter-class similarity, and domain alignment between the source and target domains, we employ multi-prototype contrastive learning between source distribution and target samples. The experiments show the effectiveness of our method on UDA benchmarks.
Label-efficient Contrastive Learning-based model for nuclei detection and classification in 3D Cardiovascular Immunofluorescent Images
Sep 08, 2023Abstract:Recently, deep learning-based methods achieved promising performance in nuclei detection and classification applications. However, training deep learning-based methods requires a large amount of pixel-wise annotated data, which is time-consuming and labor-intensive, especially in 3D images. An alternative approach is to adapt weak-annotation methods, such as labeling each nucleus with a point, but this method does not extend from 2D histopathology images (for which it was originally developed) to 3D immunofluorescent images. The reason is that 3D images contain multiple channels (z-axis) for nuclei and different markers separately, which makes training using point annotations difficult. To address this challenge, we propose the Label-efficient Contrastive learning-based (LECL) model to detect and classify various types of nuclei in 3D immunofluorescent images. Previous methods use Maximum Intensity Projection (MIP) to convert immunofluorescent images with multiple slices to 2D images, which can cause signals from different z-stacks to falsely appear associated with each other. To overcome this, we devised an Extended Maximum Intensity Projection (EMIP) approach that addresses issues using MIP. Furthermore, we performed a Supervised Contrastive Learning (SCL) approach for weakly supervised settings. We conducted experiments on cardiovascular datasets and found that our proposed framework is effective and efficient in detecting and classifying various types of nuclei in 3D immunofluorescent images.
Weakly Supervised Deep Instance Nuclei Detection using Points Annotation in 3D Cardiovascular Immunofluorescent Images
Jul 29, 2022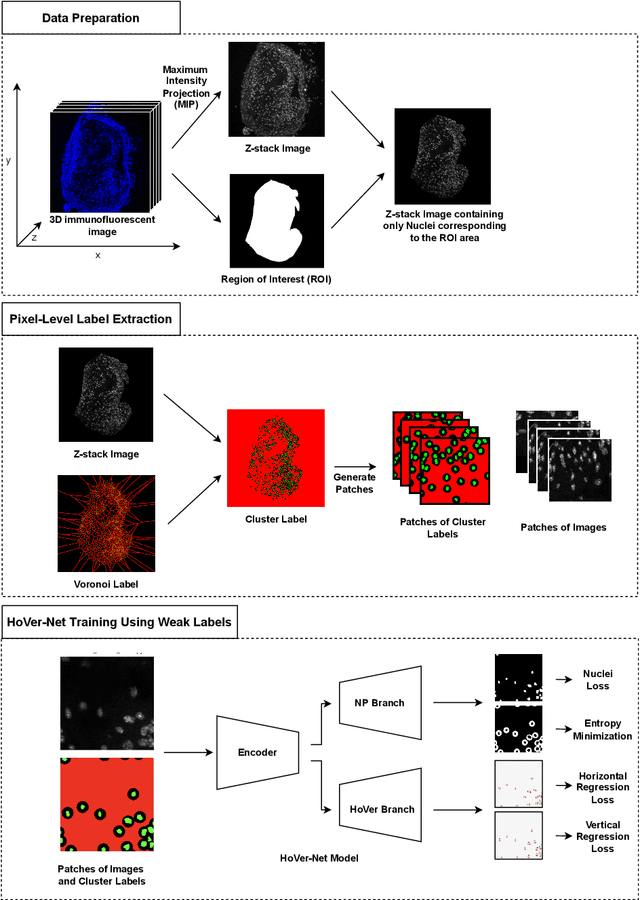

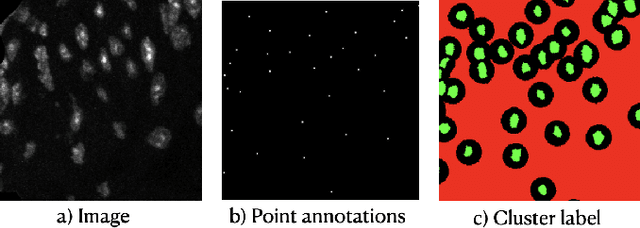
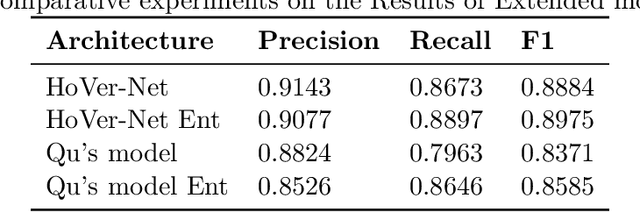
Abstract:Two major causes of death in the United States and worldwide are stroke and myocardial infarction. The underlying cause of both is thrombi released from ruptured or eroded unstable atherosclerotic plaques that occlude vessels in the heart (myocardial infarction) or the brain (stroke). Clinical studies show that plaque composition plays a more important role than lesion size in plaque rupture or erosion events. To determine the plaque composition, various cell types in 3D cardiovascular immunofluorescent images of plaque lesions are counted. However, counting these cells manually is expensive, time-consuming, and prone to human error. These challenges of manual counting motivate the need for an automated approach to localize and count the cells in images. The purpose of this study is to develop an automatic approach to accurately detect and count cells in 3D immunofluorescent images with minimal annotation effort. In this study, we used a weakly supervised learning approach to train the HoVer-Net segmentation model using point annotations to detect nuclei in fluorescent images. The advantage of using point annotations is that they require less effort as opposed to pixel-wise annotation. To train the HoVer-Net model using point annotations, we adopted a popularly used cluster labeling approach to transform point annotations into accurate binary masks of cell nuclei. Traditionally, these approaches have generated binary masks from point annotations, leaving a region around the object unlabeled (which is typically ignored during model training). However, these areas may contain important information that helps determine the boundary between cells. Therefore, we used the entropy minimization loss function in these areas to encourage the model to output more confident predictions on the unlabeled areas. Our comparison studies indicate that the HoVer-Net model trained using our weakly ...
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge