G. Venugopala Reddy
Learning Deformable 3D Graph Similarity to Track Plant Cells in Unregistered Time Lapse Images
Sep 21, 2023Abstract:Tracking of plant cells in images obtained by microscope is a challenging problem due to biological phenomena such as large number of cells, non-uniform growth of different layers of the tightly packed plant cells and cell division. Moreover, images in deeper layers of the tissue being noisy and unavoidable systemic errors inherent in the imaging process further complicates the problem. In this paper, we propose a novel learning-based method that exploits the tightly packed three-dimensional cell structure of plant cells to create a three-dimensional graph in order to perform accurate cell tracking. We further propose novel algorithms for cell division detection and effective three-dimensional registration, which improve upon the state-of-the-art algorithms. We demonstrate the efficacy of our algorithm in terms of tracking accuracy and inference-time on a benchmark dataset.
Deep Quantized Representation for Enhanced Reconstruction
Jul 29, 2021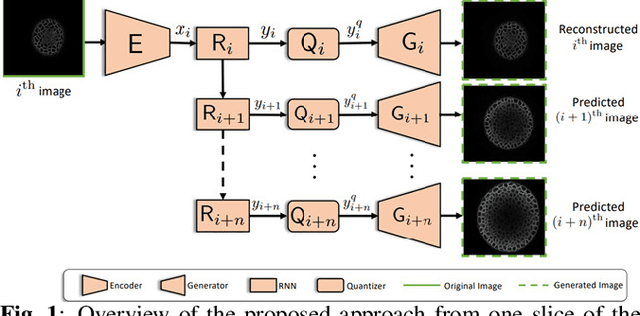
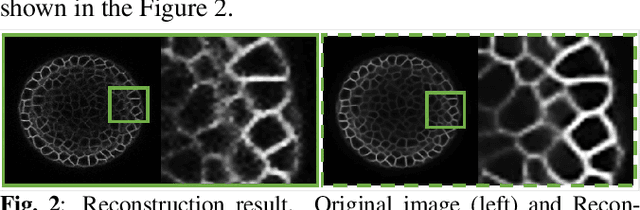
Abstract:While machine learning approaches have shown remarkable performance in biomedical image analysis, most of these methods rely on high-quality and accurate imaging data. However, collecting such data requires intensive and careful manual effort. One of the major challenges in imaging the Shoot Apical Meristem (SAM) of Arabidopsis thaliana, is that the deeper slices in the z-stack suffer from different perpetual quality-related problems like poor contrast and blurring. These quality-related issues often lead to the disposal of the painstakingly collected data with little to no control on quality while collecting the data. Therefore, it becomes necessary to employ and design techniques that can enhance the images to make them more suitable for further analysis. In this paper, we propose a data-driven Deep Quantized Latent Representation (DQLR) methodology for high-quality image reconstruction in the Shoot Apical Meristem (SAM) of Arabidopsis thaliana. Our proposed framework utilizes multiple consecutive slices in the z-stack to learn a low dimensional latent space, quantize it and subsequently perform reconstruction using the quantized representation to obtain sharper images. Experiments on a publicly available dataset validate our methodology showing promising results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge