Frank Seeliger
Parallel Capsule Networks for Classification of White Blood Cells
Sep 06, 2021



Abstract:Capsule Networks (CapsNets) is a machine learning architecture proposed to overcome some of the shortcomings of convolutional neural networks (CNNs). However, CapsNets have mainly outperformed CNNs in datasets where images are small and/or the objects to identify have minimal background noise. In this work, we present a new architecture, parallel CapsNets, which exploits the concept of branching the network to isolate certain capsules, allowing each branch to identify different entities. We applied our concept to the two current types of CapsNet architectures, studying the performance for networks with different layers of capsules. We tested our design in a public, highly unbalanced dataset of acute myeloid leukaemia images (15 classes). Our experiments showed that conventional CapsNets show similar performance than our baseline CNN (ResNeXt-50) but depict instability problems. In contrast, parallel CapsNets can outperform ResNeXt-50, is more stable, and shows better rotational invariance than both, conventional CapsNets and ResNeXt-50.
Rotaflip: A New CNN Layer for Regularization and Rotational Invariance in Medical Images
Aug 05, 2021



Abstract:Regularization in convolutional neural networks (CNNs) is usually addressed with dropout layers. However, dropout is sometimes detrimental in the convolutional part of a CNN as it simply sets to zero a percentage of pixels in the feature maps, adding unrepresentative examples during training. Here, we propose a CNN layer that performs regularization by applying random rotations of reflections to a small percentage of feature maps after every convolutional layer. We prove how this concept is beneficial for images with orientational symmetries, such as in medical images, as it provides a certain degree of rotational invariance. We tested this method in two datasets, a patch-based set of histopathology images (PatchCamelyon) to perform classification using a generic DenseNet, and a set of specular microscopy images of the corneal endothelium to perform segmentation using a tailored U-net, improving the performance in both cases.
Redesigning Fully Convolutional DenseUNets for Large Histopathology Images
Aug 05, 2021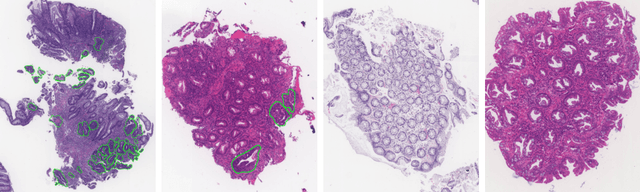
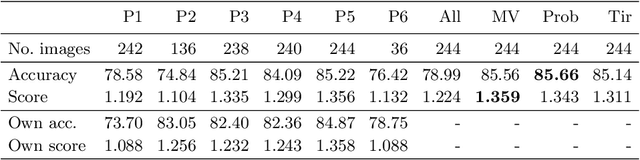
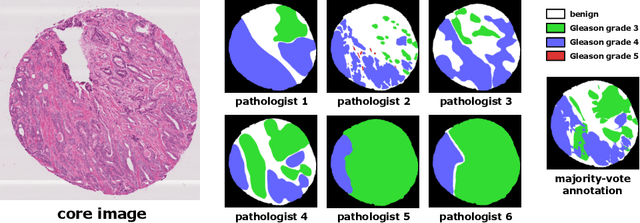
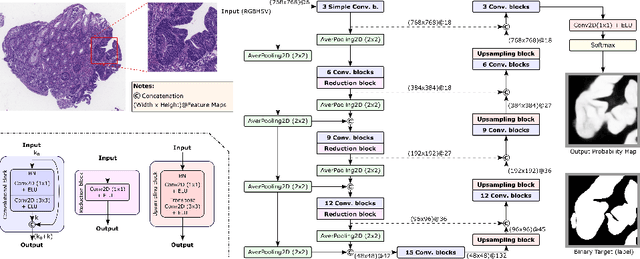
Abstract:The automated segmentation of cancer tissue in histopathology images can help clinicians to detect, diagnose, and analyze such disease. Different from other natural images used in many convolutional networks for benchmark, histopathology images can be extremely large, and the cancerous patterns can reach beyond 1000 pixels. Therefore, the well-known networks in the literature were never conceived to handle these peculiarities. In this work, we propose a Fully Convolutional DenseUNet that is particularly designed to solve histopathology problems. We evaluated our network in two public pathology datasets published as challenges in the recent MICCAI 2019: binary segmentation in colon cancer images (DigestPath2019), and multi-class segmentation in prostate cancer images (Gleason2019), achieving similar and better results than the winners of the challenges, respectively. Furthermore, we discussed some good practices in the training setup to yield the best performance and the main challenges in these histopathology datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge