Francisco J. Ribadas-Pena
Surfing the modeling of PoS taggers in low-resource scenarios
Feb 04, 2024



Abstract:The recent trend towards the application of deep structured techniques has revealed the limits of huge models in natural language processing. This has reawakened the interest in traditional machine learning algorithms, which have proved still to be competitive in certain contexts, in particular low-resource settings. In parallel, model selection has become an essential task to boost performance at reasonable cost, even more so when we talk about processes involving domains where the training and/or computational resources are scarce. Against this backdrop, we evaluate the early estimation of learning curves as a practical mechanism for selecting the most appropriate model in scenarios characterized by the use of non-deep learners in resource-lean settings. On the basis of a formal approximation model previously evaluated under conditions of wide availability of training and validation resources, we study the reliability of such an approach in a different and much more demanding operationalenvironment. Using as case study the generation of PoS taggers for Galician, a language belonging to the Western Ibero-Romance group, the experimental results are consistent with our expectations.
* 17 papes, 5 figures
Improving Large-Scale k-Nearest Neighbor Text Categorization with Label Autoencoders
Feb 03, 2024Abstract:In this paper, we introduce a multi-label lazy learning approach to deal with automatic semantic indexing in large document collections in the presence of complex and structured label vocabularies with high inter-label correlation. The proposed method is an evolution of the traditional k-Nearest Neighbors algorithm which uses a large autoencoder trained to map the large label space to a reduced size latent space and to regenerate the predicted labels from this latent space. We have evaluated our proposal in a large portion of the MEDLINE biomedical document collection which uses the Medical Subject Headings (MeSH) thesaurus as a controlled vocabulary. In our experiments we propose and evaluate several document representation approaches and different label autoencoder configurations.
* 22 pages, 4 figures
CoLe and LYS at BioASQ MESINESP8 Task: similarity based descriptor assignment in Spanish
Feb 02, 2024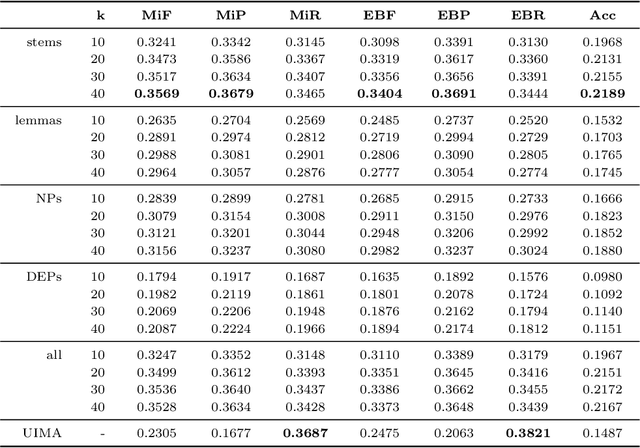

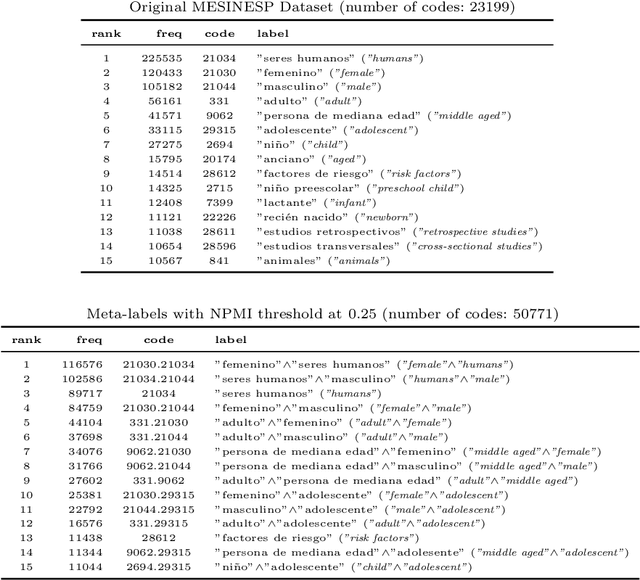
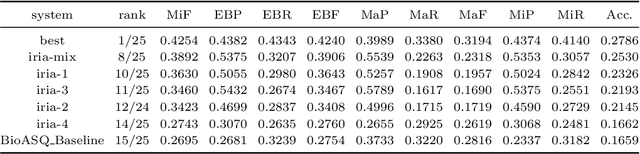
Abstract:In this paper, we describe our participation in the MESINESP Task of the BioASQ biomedical semantic indexing challenge. The participating system follows an approach based solely on conventional information retrieval tools. We have evaluated various alternatives for extracting index terms from IBECS/LILACS documents in order to be stored in an Apache Lucene index. Those indexed representations are queried using the contents of the article to be annotated and a ranked list of candidate labels is created from the retrieved documents. We also have evaluated a sort of limited Label Powerset approach which creates meta-labels joining pairs of DeCS labels with high co-occurrence scores, and an alternative method based on label profile matching. Results obtained in official runs seem to confirm the suitability of this approach for languages like Spanish.
* Accepted at the 8th BioASQ Workshop at the 11th Conference and Labs of the Evaluation Forum (CLEF) 2020. 11 pages
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge