Fatemeh Esfahani
Multi-Stage Graph Peeling Algorithm for Probabilistic Core Decomposition
Aug 13, 2021



Abstract:Mining dense subgraphs where vertices connect closely with each other is a common task when analyzing graphs. A very popular notion in subgraph analysis is core decomposition. Recently, Esfahani et al. presented a probabilistic core decomposition algorithm based on graph peeling and Central Limit Theorem (CLT) that is capable of handling very large graphs. Their proposed peeling algorithm (PA) starts from the lowest degree vertices and recursively deletes these vertices, assigning core numbers, and updating the degree of neighbour vertices until it reached the maximum core. However, in many applications, particularly in biology, more valuable information can be obtained from dense sub-communities and we are not interested in small cores where vertices do not interact much with others. To make the previous PA focus more on dense subgraphs, we propose a multi-stage graph peeling algorithm (M-PA) that has a two-stage data screening procedure added before the previous PA. After removing vertices from the graph based on the user-defined thresholds, we can reduce the graph complexity largely and without affecting the vertices in subgraphs that we are interested in. We show that M-PA is more efficient than the previous PA and with the properly set filtering threshold, can produce very similar if not identical dense subgraphs to the previous PA (in terms of graph density and clustering coefficient).
Lightweight Combinational Machine Learning Algorithm for Sorting Canine Torso Radiographs
Feb 22, 2021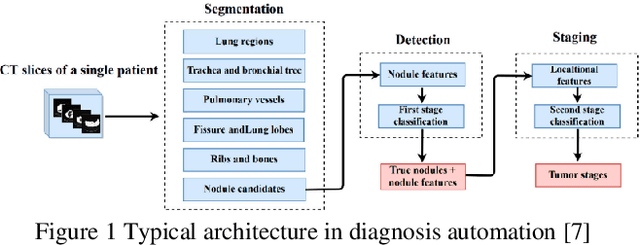
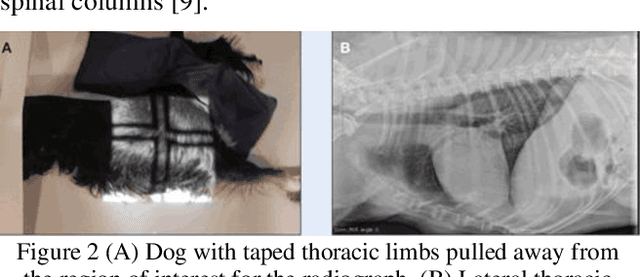
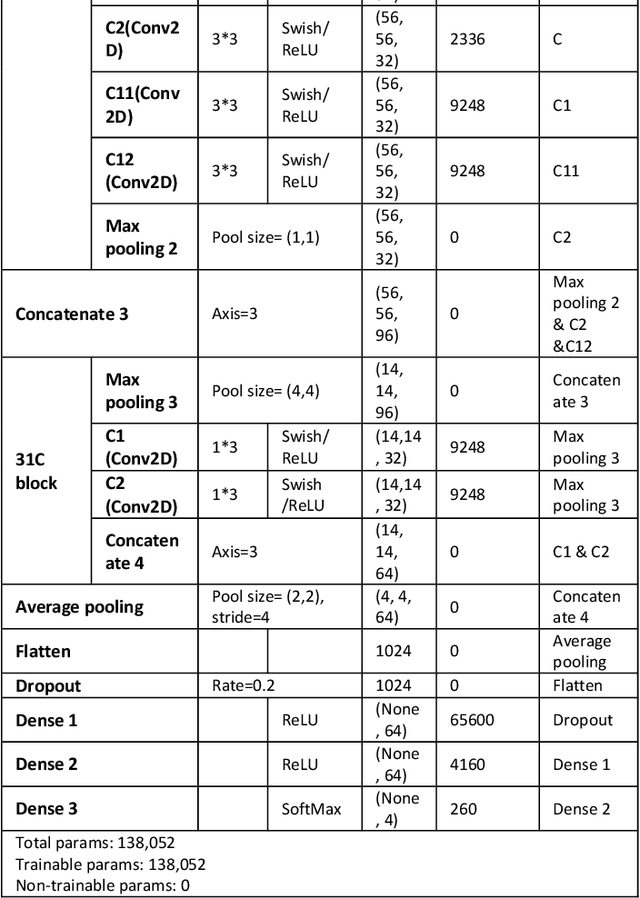

Abstract:The veterinary field lacks automation in contrast to the tremendous technological advances made in the human medical field. Implementation of machine learning technology can shorten any step of the automation process. This paper explores these core concepts and starts with automation in sorting radiographs for canines by view and anatomy. This is achieved by developing a new lightweight algorithm inspired by AlexNet, Inception, and SqueezeNet. The proposed module proves to be lighter than SqueezeNet while maintaining accuracy higher than that of AlexNet, ResNet, DenseNet, and SqueezeNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge