Farnaz H. Foomani
Machine learning techniques to identify antibiotic resistance in patients diagnosed with various skin and soft tissue infections
Feb 28, 2022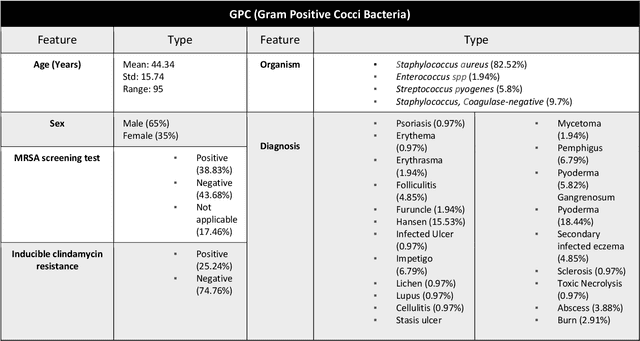
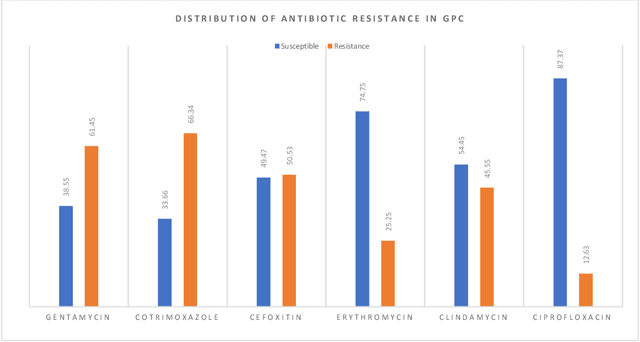
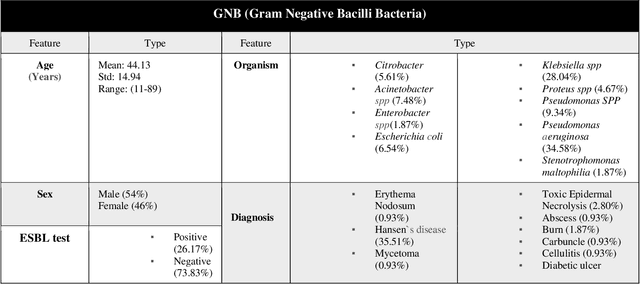
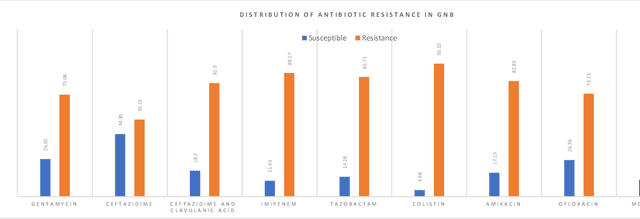
Abstract:Skin and soft tissue infections (SSTIs) are among the most frequently observed diseases in ambulatory and hospital settings. Resistance of diverse bacterial pathogens to antibiotics is a significant cause of severe SSTIs, and treatment failure results in morbidity, mortality, and increased cost of hospitalization. Therefore, antimicrobial surveillance is essential to predict antibiotic resistance trends and monitor the results of medical interventions. To address this, we developed machine learning (ML) models (deep and conventional algorithms) to predict antimicrobial resistance using antibiotic susceptibility testing (ABST) data collected from patients clinically diagnosed with primary and secondary pyoderma over a period of one year. We trained an individual ML algorithm on each antimicrobial family to determine whether a Gram-Positive Cocci (GPC) or Gram-Negative Bacilli (GNB) bacteria will resist the corresponding antibiotic. For this purpose, clinical and demographic features from the patient and data from ABST were employed in training. We achieved an Area Under the Curve (AUC) of 0.68-0.98 in GPC and 0.56-0.93 in GNB bacteria, depending on the antimicrobial family. We also conducted a correlation analysis to determine the linear relationship between each feature and antimicrobial families in different bacteria. ML techniques suggest that a predictable nonlinear relationship exists between patients' clinical-demographic characteristics and antibiotic resistance; however, the accuracy of this prediction depends on the type of the antimicrobial family.
Synthesizing time-series wound prognosis factors from electronic medical records using generative adversarial networks
May 03, 2021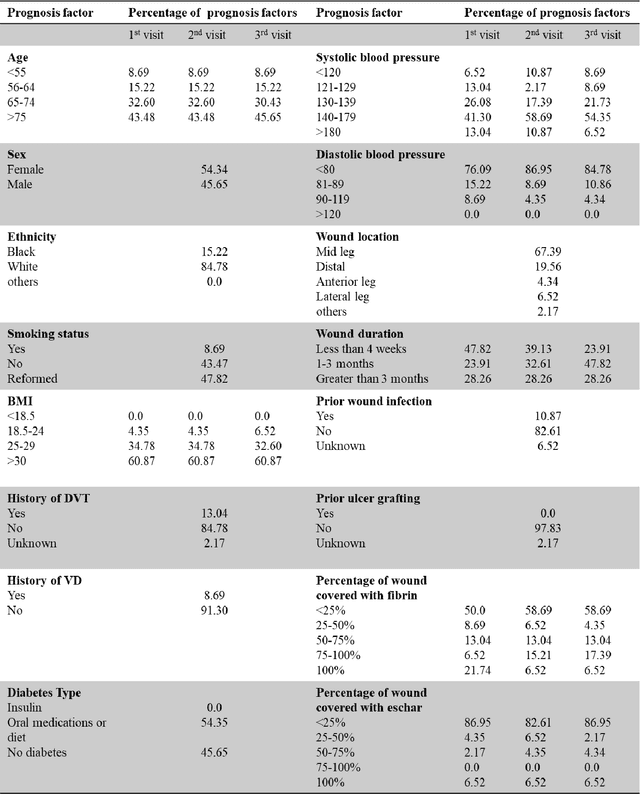
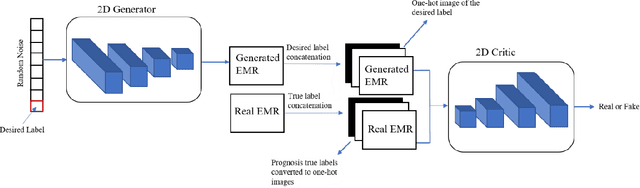
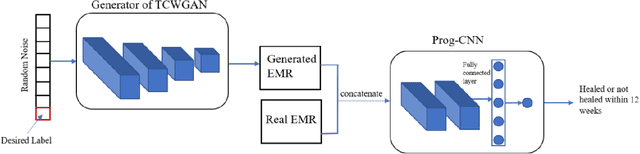
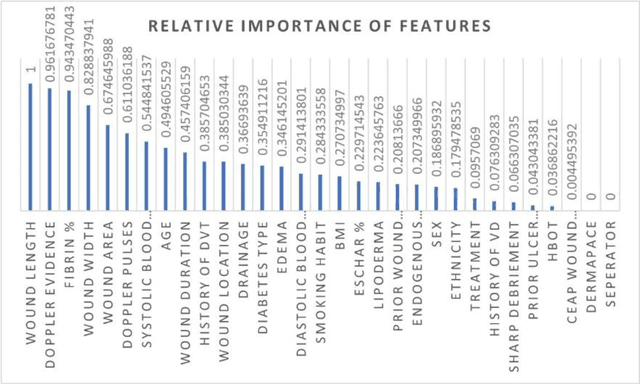
Abstract:Wound prognostic models not only provide an estimate of wound healing time to motivate patients to follow up their treatments but also can help clinicians to decide whether to use a standard care or adjuvant therapies and to assist them with designing clinical trials. However, collecting prognosis factors from Electronic Medical Records (EMR) of patients is challenging due to privacy, sensitivity, and confidentiality. In this study, we developed time series medical generative adversarial networks (GANs) to generate synthetic wound prognosis factors using very limited information collected during routine care in a specialized wound care facility. The generated prognosis variables are used in developing a predictive model for chronic wound healing trajectory. Our novel medical GAN can produce both continuous and categorical features from EMR. Moreover, we applied temporal information to our model by considering data collected from the weekly follow-ups of patients. Conditional training strategies were utilized to enhance training and generate classified data in terms of healing or non-healing. The ability of the proposed model to generate realistic EMR data was evaluated by TSTR (test on the synthetic, train on the real), discriminative accuracy, and visualization. We utilized samples generated by our proposed GAN in training a prognosis model to demonstrate its real-life application. Using the generated samples in training predictive models improved the classification accuracy by 6.66-10.01% compared to the previous EMR-GAN. Additionally, the suggested prognosis classifier has achieved the area under the curve (AUC) of 0.975, 0.968, and 0.849 when training the network using data from the first three visits, first two visits, and first visit, respectively. These results indicate a significant improvement in wound healing prediction compared to the previous prognosis models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge