Ewelina Weglarz-Tomczak
De Novo Drug Design with Joint Transformers
Oct 03, 2023Abstract:De novo drug design requires simultaneously generating novel molecules outside of training data and predicting their target properties, making it a hard task for generative models. To address this, we propose Joint Transformer that combines a Transformer decoder, a Transformer encoder, and a predictor in a joint generative model with shared weights. We show that training the model with a penalized log-likelihood objective results in state-of-the-art performance in molecule generation, while decreasing the prediction error on newly sampled molecules, as compared to a fine-tuned decoder-only Transformer, by 42%. Finally, we propose a probabilistic black-box optimization algorithm that employs Joint Transformer to generate novel molecules with improved target properties, as compared to the training data, outperforming other SMILES-based optimization methods in de novo drug design.
Population-based Optimization for Kinetic Parameter Identification in Glycolytic Pathway in Saccharomyces cerevisiae
Sep 19, 2020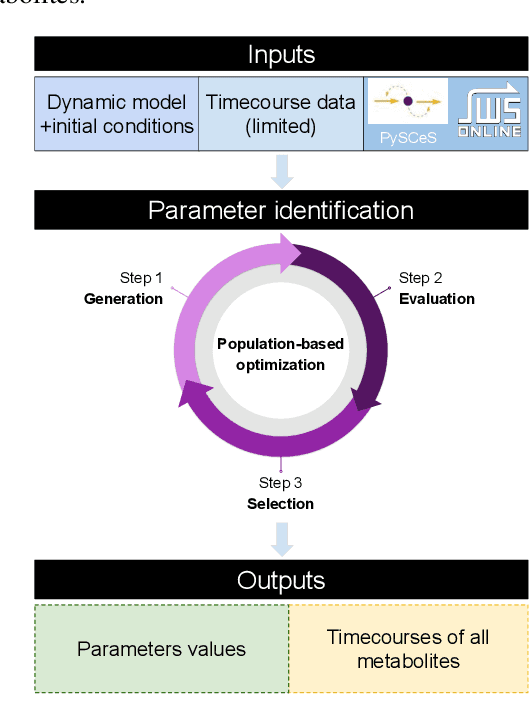
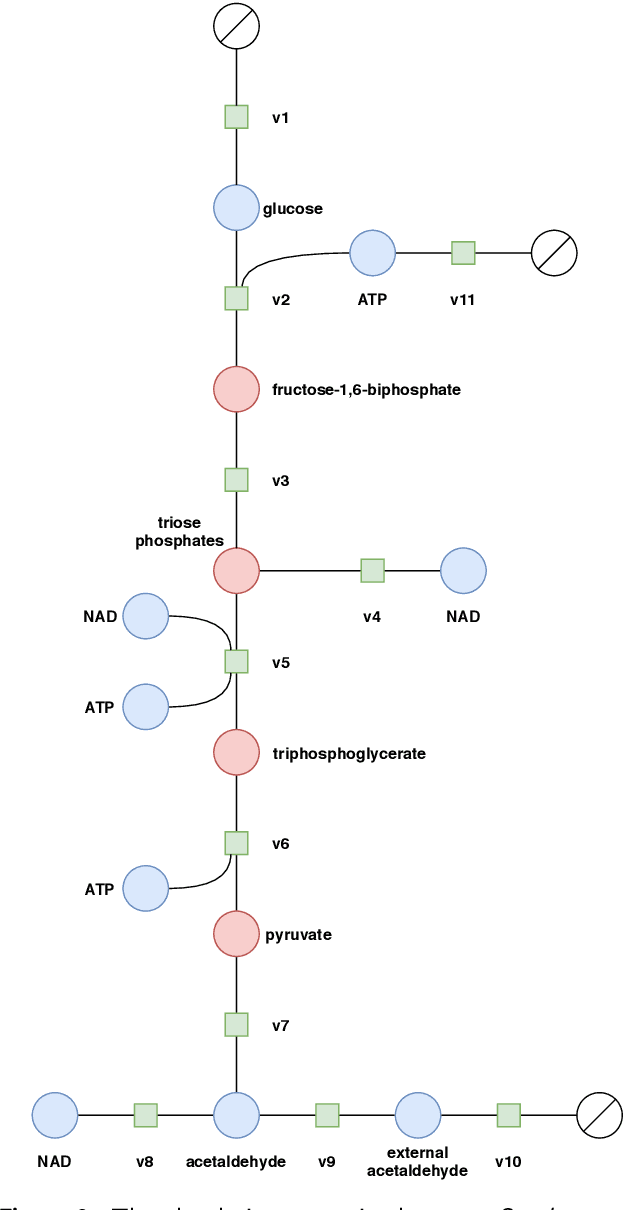

Abstract:Models in systems biology are mathematical descriptions of biological processes that are used to answer questions and gain a better understanding of biological phenomena. Dynamic models represent the network through rates of the production and consumption for the individual species. The ordinary differential equations that describe rates of the reactions in the model include a set of parameters. The parameters are important quantities to understand and analyze biological systems. Moreover, the perturbation of the kinetic parameters are correlated with upregulation of the system by cell-intrinsic and cell-extrinsic factors, including mutations and the environment changes. Here, we aim at using well-established models of biological pathways to identify parameter values and point their potential perturbation/deviation. We present our population-based optimization framework that is able to identify kinetic parameters in the dynamic model based on only input and output data (i.e., timecourses of selected metabolites). Our approach can deal with the identification of the non-measurable parameters as well as with discovering deviation of the parameters. We present our proposed optimization framework on the example of the well-studied glycolytic pathway in Saccharomyces cerevisiae.
Differential Evolution with Reversible Linear Transformations
Feb 07, 2020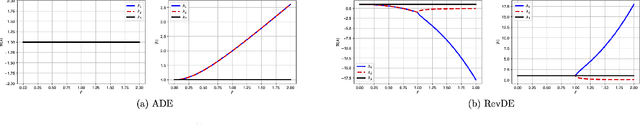

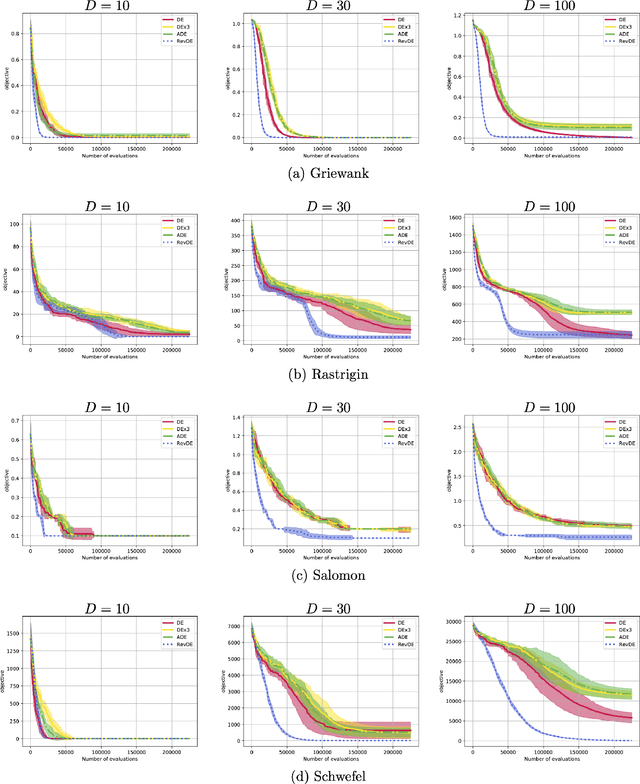
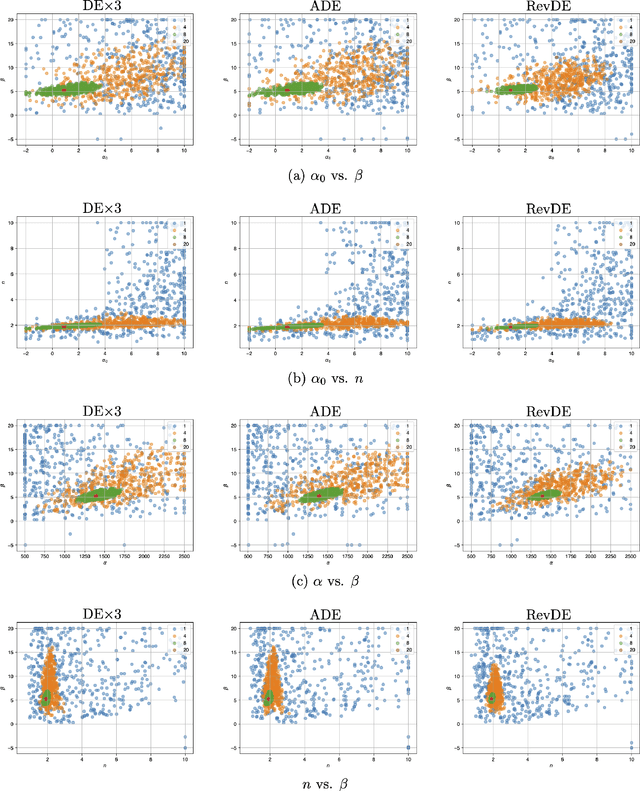
Abstract:Differential evolution (DE) is a well-known type of evolutionary algorithms (EA). Similarly to other EA variants it can suffer from small populations and loose diversity too quickly. This paper presents a new approach to mitigate this issue: We propose to generate new candidate solutions by utilizing reversible linear transformation applied to a triplet of solutions from the population. In other words, the population is enlarged by using newly generated individuals without evaluating their fitness. We assess our methods on three problems: (i) benchmark function optimization, (ii) discovering parameter values of the gene repressilator system, (iii) learning neural networks. The empirical results indicate that the proposed approach outperforms vanilla DE and a version of DE with applying differential mutation three times on all testbeds.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge