Eric W. Klee
Daphne
Deep Phenotyping of Non-Alcoholic Fatty Liver Disease Patients with Genetic Factors for Insights into the Complex Disease
Nov 13, 2023Abstract:Non-alcoholic fatty liver disease (NAFLD) is a prevalent chronic liver disorder characterized by the excessive accumulation of fat in the liver in individuals who do not consume significant amounts of alcohol, including risk factors like obesity, insulin resistance, type 2 diabetes, etc. We aim to identify subgroups of NAFLD patients based on demographic, clinical, and genetic characteristics for precision medicine. The genomic and phenotypic data (3,408 cases and 4,739 controls) for this study were gathered from participants in Mayo Clinic Tapestry Study (IRB#19-000001) and their electric health records, including their demographic, clinical, and comorbidity data, and the genotype information through whole exome sequencing performed at Helix using the Exome+$^\circledR$ Assay according to standard procedure (www$.$helix$.$com). Factors highly relevant to NAFLD were determined by the chi-square test and stepwise backward-forward regression model. Latent class analysis (LCA) was performed on NAFLD cases using significant indicator variables to identify subgroups. The optimal clustering revealed 5 latent subgroups from 2,013 NAFLD patients (mean age 60.6 years and 62.1% women), while a polygenic risk score based on 6 single-nucleotide polymorphism (SNP) variants and disease outcomes were used to analyze the subgroups. The groups are characterized by metabolic syndrome, obesity, different comorbidities, psychoneurological factors, and genetic factors. Odds ratios were utilized to compare the risk of complex diseases, such as fibrosis, cirrhosis, and hepatocellular carcinoma (HCC), as well as liver failure between the clusters. Cluster 2 has a significantly higher complex disease outcome compared to other clusters. Keywords: Fatty liver disease; Polygenic risk score; Precision medicine; Deep phenotyping; NAFLD comorbidities; Latent class analysis.
Universally Rank Consistent Ordinal Regression in Neural Networks
Oct 14, 2021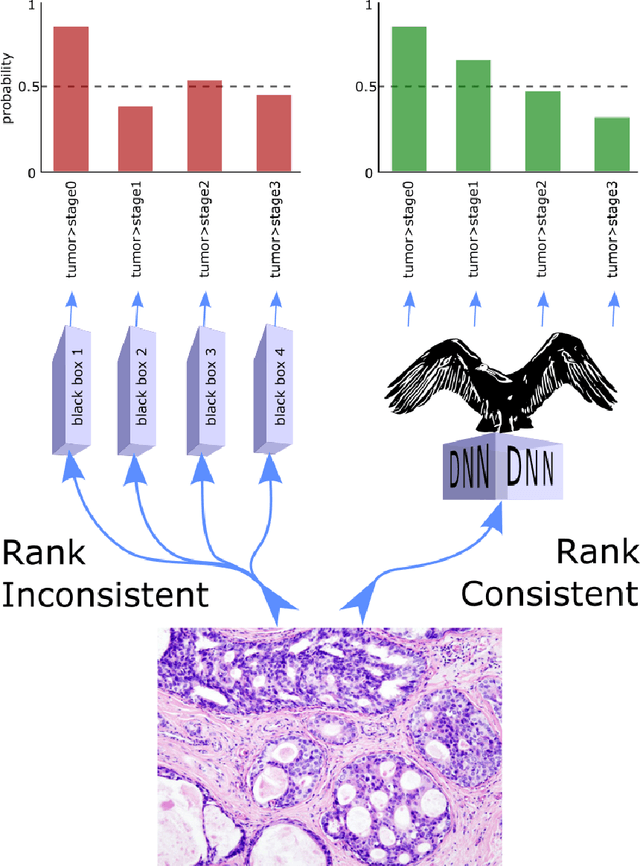
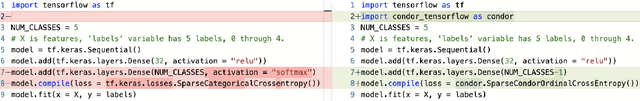
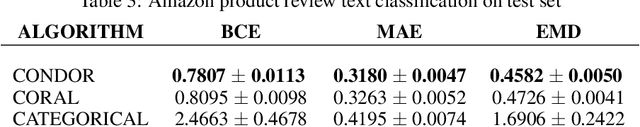
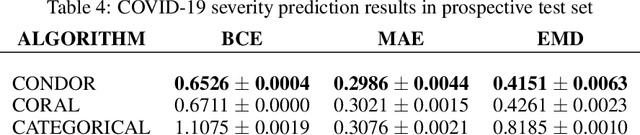
Abstract:Despite the pervasiveness of ordinal labels in supervised learning, it remains common practice in deep learning to treat such problems as categorical classification using the categorical cross entropy loss. Recent methods attempting to address this issue while respecting the ordinal structure of the labels have resorted to converting ordinal regression into a series of extended binary classification subtasks. However, the adoption of such methods remains inconsistent due to theoretical and practical limitations. Here we address these limitations by demonstrating that the subtask probabilities form a Markov chain. We show how to straightforwardly modify neural network architectures to exploit this fact and thereby constrain predictions to be universally rank consistent. We furthermore prove that all rank consistent solutions can be represented within this formulation. Using diverse benchmarks and the real-world application of a specialized recurrent neural network for COVID-19 prognosis, we demonstrate the practical superiority of this method versus the current state-of-the-art. The method is open sourced as user-friendly PyTorch and TensorFlow packages.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge