Domingo Alcaraz-Segura
Department of Botany, Faculty of Science, University of Granada, Granada, Spain
Deep Learning for blind spectral unmixing of LULC classes with MODIS multispectral time series and ancillary data
Oct 11, 2023Abstract:Remotely sensed data are dominated by mixed Land Use and Land Cover (LULC) types. Spectral unmixing is a technique to extract information from mixed pixels into their constituent LULC types and corresponding abundance fractions. Traditionally, solving this task has relied on either classical methods that require prior knowledge of endmembers or machine learning methods that avoid explicit endmembers calculation, also known as blind spectral unmixing (BSU). Most BSU studies based on Deep Learning (DL) focus on one time-step hyperspectral data, yet its acquisition remains quite costly compared with multispectral data. To our knowledge, here we provide the first study on BSU of LULC classes using multispectral time series data with DL models. We further boost the performance of a Long-Short Term Memory (LSTM)-based model by incorporating geographic plus topographic (geo-topographic) and climatic ancillary information. Our experiments show that combining spectral-temporal input data together with geo-topographic and climatic information substantially improves the abundance estimation of LULC classes in mixed pixels. To carry out this study, we built a new labeled dataset of the region of Andalusia (Spain) with monthly multispectral time series of pixels for the year 2013 from MODIS at 460m resolution, for two hierarchical levels of LULC classes, named Andalusia MultiSpectral MultiTemporal Unmixing (Andalusia-MSMTU). This dataset provides, at the pixel level, a multispectral time series plus ancillary information annotated with the abundance of each LULC class inside each pixel. The dataset and code are available to the public.
Deep-Learning Convolutional Neural Networks for scattered shrub detection with Google Earth Imagery
Jun 03, 2017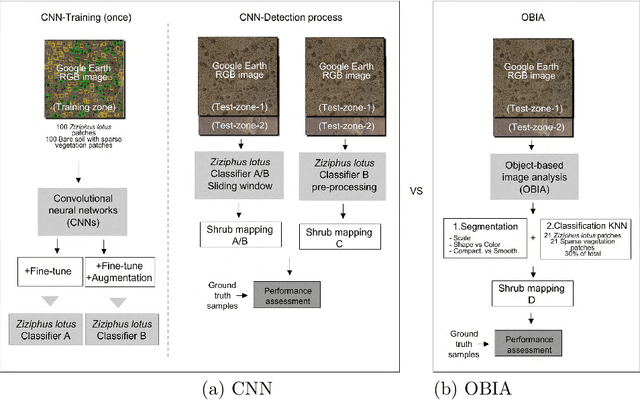
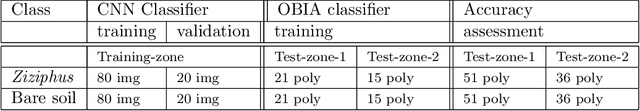

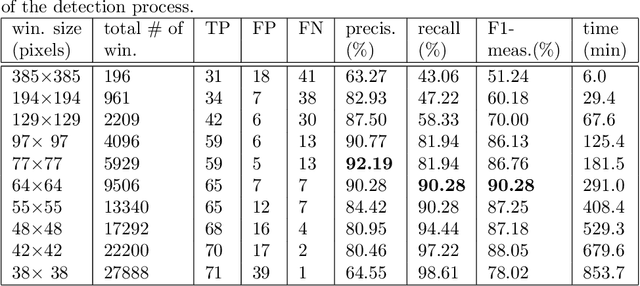
Abstract:There is a growing demand for accurate high-resolution land cover maps in many fields, e.g., in land-use planning and biodiversity conservation. Developing such maps has been performed using Object-Based Image Analysis (OBIA) methods, which usually reach good accuracies, but require a high human supervision and the best configuration for one image can hardly be extrapolated to a different image. Recently, the deep learning Convolutional Neural Networks (CNNs) have shown outstanding results in object recognition in the field of computer vision. However, they have not been fully explored yet in land cover mapping for detecting species of high biodiversity conservation interest. This paper analyzes the potential of CNNs-based methods for plant species detection using free high-resolution Google Earth T M images and provides an objective comparison with the state-of-the-art OBIA-methods. We consider as case study the detection of Ziziphus lotus shrubs, which are protected as a priority habitat under the European Union Habitats Directive. According to our results, compared to OBIA-based methods, the proposed CNN-based detection model, in combination with data-augmentation, transfer learning and pre-processing, achieves higher performance with less human intervention and the knowledge it acquires in the first image can be transferred to other images, which makes the detection process very fast. The provided methodology can be systematically reproduced for other species detection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge