Dmitrij Rappoport
Space-Filling Curves as a Novel Crystal Structure Representation for Machine Learning Models
Aug 19, 2016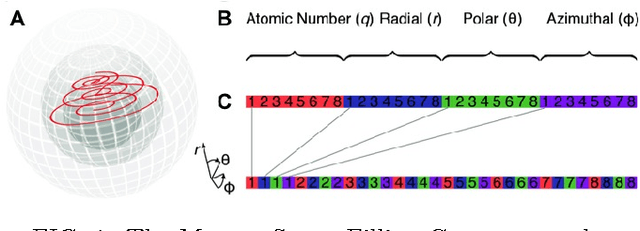
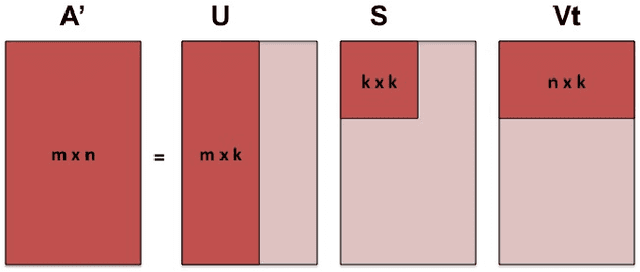
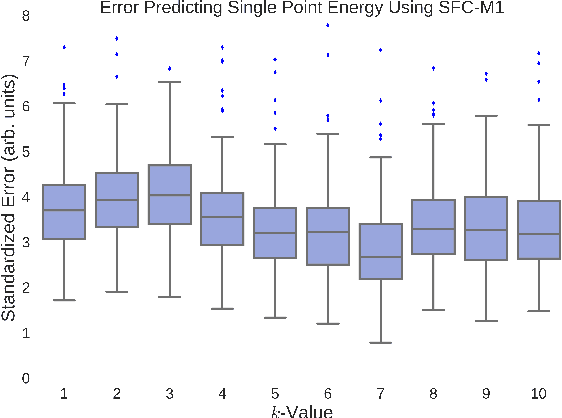
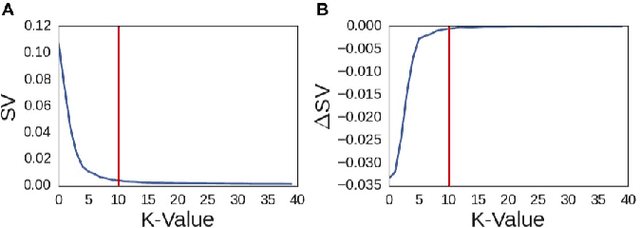
Abstract:A fundamental problem in applying machine learning techniques for chemical problems is to find suitable representations for molecular and crystal structures. While the structure representations based on atom connectivities are prevalent for molecules, two-dimensional descriptors are not suitable for describing molecular crystals. In this work, we introduce the SFC-M family of feature representations, which are based on Morton space-filling curves, as an alternative means of representing crystal structures. Latent Semantic Indexing (LSI) was employed in a novel setting to reduce sparsity of feature representations. The quality of the SFC-M representations were assessed by using them in combination with artificial neural networks to predict Density Functional Theory (DFT) single point, Ewald summed, lattice, and many-body dispersion energies of 839 organic molecular crystal unit cells from the Cambridge Structural Database that consist of the elements C, H, N, and O. Promising initial results suggest that the SFC-M representations merit further exploration to improve its ability to predict solid-state properties of organic crystal structures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge