Der-Chen Chang
Self-supervised Representation Learning on Electronic Health Records with Graph Kernel Infomax
Sep 01, 2022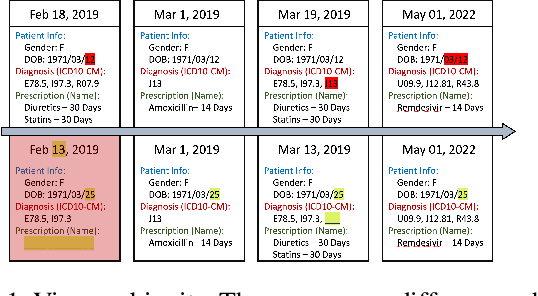
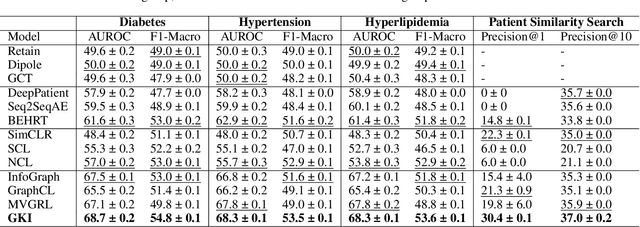
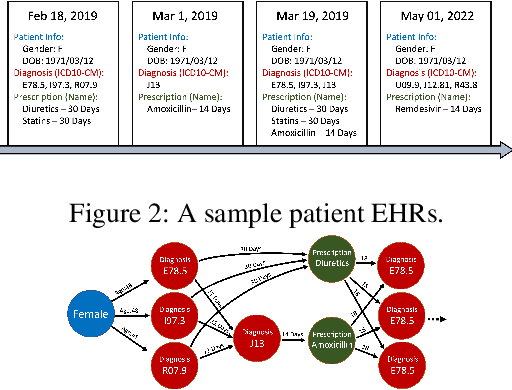

Abstract:Learning Electronic Health Records (EHRs) representation is a preeminent yet under-discovered research topic. It benefits various clinical decision support applications, e.g., medication outcome prediction or patient similarity search. Current approaches focus on task-specific label supervision on vectorized sequential EHR, which is not applicable to large-scale unsupervised scenarios. Recently, contrastive learning shows great success on self-supervised representation learning problems. However, complex temporality often degrades the performance. We propose Graph Kernel Infomax, a self-supervised graph kernel learning approach on the graphical representation of EHR, to overcome the previous problems. Unlike the state-of-the-art, we do not change the graph structure to construct augmented views. Instead, we use Kernel Subspace Augmentation to embed nodes into two geometrically different manifold views. The entire framework is trained by contrasting nodes and graph representations on those two manifold views through the commonly used contrastive objectives. Empirically, using publicly available benchmark EHR datasets, our approach yields performance on clinical downstream tasks that exceeds the state-of-the-art. Theoretically, the variation on distance metrics naturally creates different views as data augmentation without changing graph structures.
The Analysis from Nonlinear Distance Metric to Kernel-based Drug Prescription Prediction System
Feb 24, 2021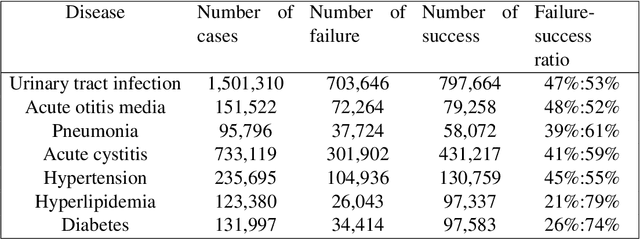
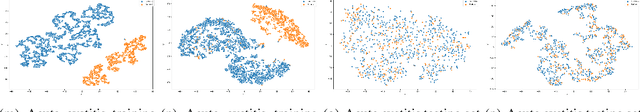
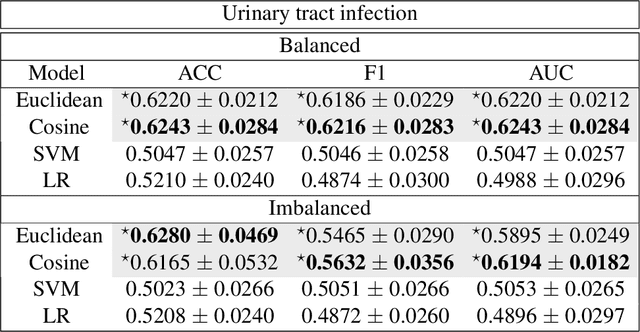
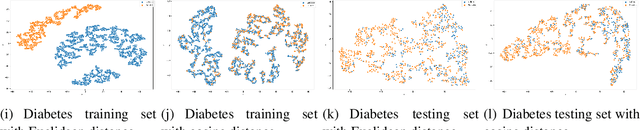
Abstract:Distance metrics and their nonlinear variant play a crucial role in machine learning based real-world problem solving. We demonstrated how Euclidean and cosine distance measures differ not only theoretically but also in real-world medical application, namely, outcome prediction of drug prescription. Euclidean distance exhibits favorable properties in the local geometry problem. To this regard, Euclidean distance can be applied under short-term disease with low-variation outcome observation. Moreover, when presenting to highly variant chronic disease, it is preferable to use cosine distance. These different geometric properties lead to different submanifolds in the original embedded space, and hence, to different optimizing nonlinear kernel embedding frameworks. We first established the geometric properties that we needed in these frameworks. From these properties interpreted their differences in certain perspectives. Our evaluation on real-world, large-scale electronic health records and embedding space visualization empirically validated our approach.
Cross-Global Attention Graph Kernel Network Prediction of Drug Prescription
Aug 04, 2020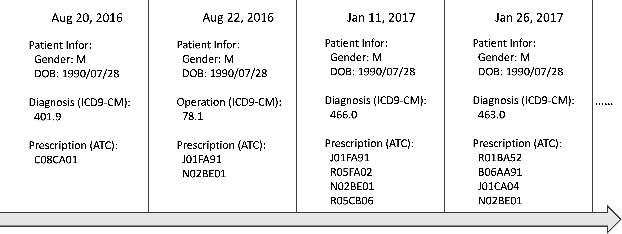
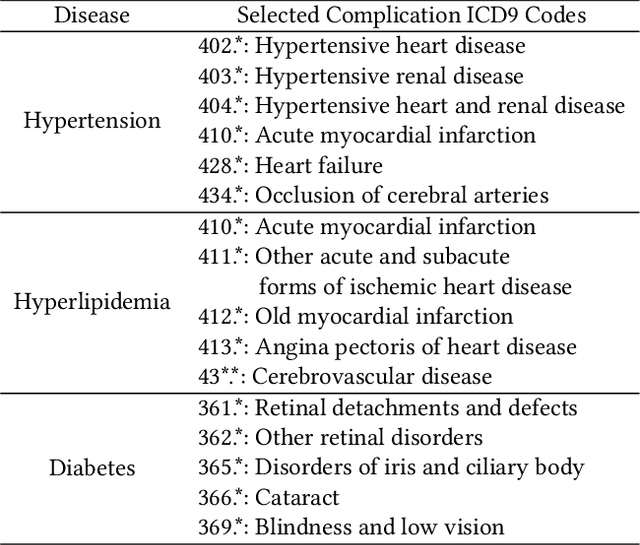
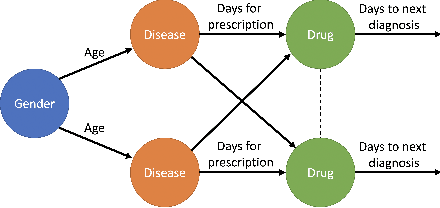

Abstract:We present an end-to-end, interpretable, deep-learning architecture to learn a graph kernel that predicts the outcome of chronic disease drug prescription. This is achieved through a deep metric learning collaborative with a Support Vector Machine objective using a graphical representation of Electronic Health Records. We formulate the predictive model as a binary graph classification problem with an adaptive learned graph kernel through novel cross-global attention node matching between patient graphs, simultaneously computing on multiple graphs without training pair or triplet generation. Results using the Taiwanese National Health Insurance Research Database demonstrate that our approach outperforms current start-of-the-art models both in terms of accuracy and interpretability.
* ACM-BCB 2020 (Full paper)
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge