Nairen Cao
Breaking 3-Factor Approximation for Correlation Clustering in Polylogarithmic Rounds
Jul 13, 2023Abstract:In this paper, we study parallel algorithms for the correlation clustering problem, where every pair of two different entities is labeled with similar or dissimilar. The goal is to partition the entities into clusters to minimize the number of disagreements with the labels. Currently, all efficient parallel algorithms have an approximation ratio of at least 3. In comparison with the $1.994+\epsilon$ ratio achieved by polynomial-time sequential algorithms [CLN22], a significant gap exists. We propose the first poly-logarithmic depth parallel algorithm that achieves a better approximation ratio than 3. Specifically, our algorithm computes a $(2.4+\epsilon)$-approximate solution and uses $\tilde{O}(m^{1.5})$ work. Additionally, it can be translated into a $\tilde{O}(m^{1.5})$-time sequential algorithm and a poly-logarithmic rounds sublinear-memory MPC algorithm with $\tilde{O}(m^{1.5})$ total memory. Our approach is inspired by Awerbuch, Khandekar, and Rao's [AKR12] length-constrained multi-commodity flow algorithm, where we develop an efficient parallel algorithm to solve a truncated correlation clustering linear program of Charikar, Guruswami, and Wirth [CGW05]. Then we show the solution of the truncated linear program can be rounded with a factor of at most 2.4 loss by using the framework of [CMSY15]. Such a rounding framework can then be implemented using parallel pivot-based approaches.
Self-supervised Representation Learning on Electronic Health Records with Graph Kernel Infomax
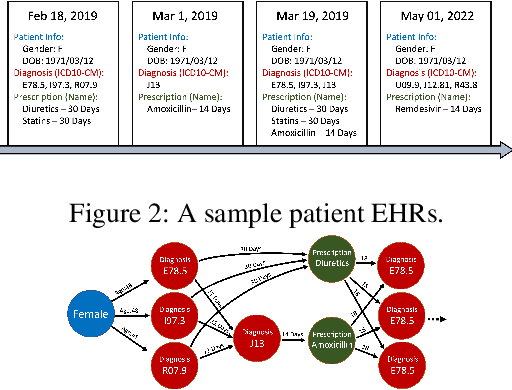
Sep 01, 2022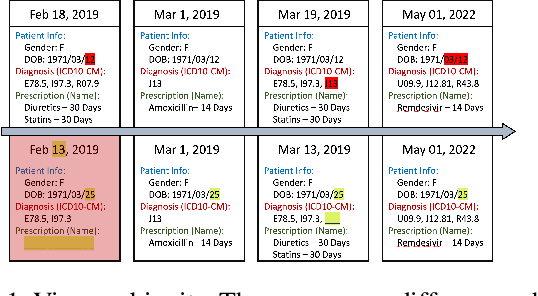
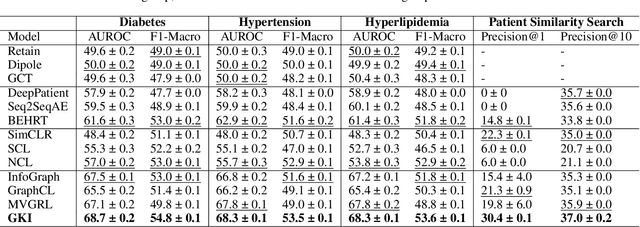

Abstract:Learning Electronic Health Records (EHRs) representation is a preeminent yet under-discovered research topic. It benefits various clinical decision support applications, e.g., medication outcome prediction or patient similarity search. Current approaches focus on task-specific label supervision on vectorized sequential EHR, which is not applicable to large-scale unsupervised scenarios. Recently, contrastive learning shows great success on self-supervised representation learning problems. However, complex temporality often degrades the performance. We propose Graph Kernel Infomax, a self-supervised graph kernel learning approach on the graphical representation of EHR, to overcome the previous problems. Unlike the state-of-the-art, we do not change the graph structure to construct augmented views. Instead, we use Kernel Subspace Augmentation to embed nodes into two geometrically different manifold views. The entire framework is trained by contrasting nodes and graph representations on those two manifold views through the commonly used contrastive objectives. Empirically, using publicly available benchmark EHR datasets, our approach yields performance on clinical downstream tasks that exceeds the state-of-the-art. Theoretically, the variation on distance metrics naturally creates different views as data augmentation without changing graph structures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge