Dehua Peng
Interpreting the Curse of Dimensionality from Distance Concentration and Manifold Effect
Jan 07, 2024Abstract:The characteristics of data like distribution and heterogeneity, become more complex and counterintuitive as the dimensionality increases. This phenomenon is known as curse of dimensionality, where common patterns and relationships (e.g., internal and boundary pattern) that hold in low-dimensional space may be invalid in higher-dimensional space. It leads to a decreasing performance for the regression, classification or clustering models or algorithms. Curse of dimensionality can be attributed to many causes. In this paper, we first summarize five challenges associated with manipulating high-dimensional data, and explains the potential causes for the failure of regression, classification or clustering tasks. Subsequently, we delve into two major causes of the curse of dimensionality, distance concentration and manifold effect, by performing theoretical and empirical analyses. The results demonstrate that nearest neighbor search (NNS) using three typical distance measurements, Minkowski distance, Chebyshev distance, and cosine distance, becomes meaningless as the dimensionality increases. Meanwhile, the data incorporates more redundant features, and the variance contribution of principal component analysis (PCA) is skewed towards a few dimensions. By interpreting the causes of the curse of dimensionality, we can better understand the limitations of current models and algorithms, and drive to improve the performance of data analysis and machine learning tasks in high-dimensional space.
Scalable manifold learning by uniform landmark sampling and constrained locally linear embedding
Jan 05, 2024Abstract:As a pivotal approach in machine learning and data science, manifold learning aims to uncover the intrinsic low-dimensional structure within complex nonlinear manifolds in high-dimensional space. By exploiting the manifold hypothesis, various techniques for nonlinear dimension reduction have been developed to facilitate visualization, classification, clustering, and gaining key insights. Although existing manifold learning methods have achieved remarkable successes, they still suffer from extensive distortions incurred in the global structure, which hinders the understanding of underlying patterns. Scalability issues also limit their applicability for handling large-scale data. Here, we propose a scalable manifold learning (scML) method that can manipulate large-scale and high-dimensional data in an efficient manner. It starts by seeking a set of landmarks to construct the low-dimensional skeleton of the entire data, and then incorporates the non-landmarks into the learned space based on the constrained locally linear embedding (CLLE). We empirically validated the effectiveness of scML on synthetic datasets and real-world benchmarks of different types, and applied it to analyze the single-cell transcriptomics and detect anomalies in electrocardiogram (ECG) signals. scML scales well with increasing data sizes and embedding dimensions, and exhibits promising performance in preserving the global structure. The experiments demonstrate notable robustness in embedding quality as the sample rate decreases.
A Robust and Efficient Boundary Point Detection Method by Measuring Local Direction Dispersion
Dec 07, 2023Abstract:Boundary points pose a significant challenge for machine learning tasks, including classification, clustering, and dimensionality reduction. Due to the similarity of features, boundary areas can result in mixed-up classes or clusters, leading to a crowding problem in dimensionality reduction. To address this challenge, numerous boundary point detection methods have been developed, but they are insufficiently to accurately and efficiently identify the boundary points in non-convex structures and high-dimensional manifolds. In this work, we propose a robust and efficient method for detecting boundary points using Local Direction Dispersion (LoDD). LoDD considers that internal points are surrounded by neighboring points in all directions, while neighboring points of a boundary point tend to be distributed only in a certain directional range. LoDD adopts a density-independent K-Nearest Neighbors (KNN) method to determine neighboring points, and defines a statistic-based metric using the eigenvalues of the covariance matrix of KNN coordinates to measure the centrality of a query point. We demonstrated the validity of LoDD on five synthetic datasets (2-D and 3-D) and ten real-world benchmarks, and tested its clustering performance by equipping with two typical clustering methods, K-means and Ncut. Our results show that LoDD achieves promising and robust detection accuracy in a time-efficient manner.
MeanCut: A Greedy-Optimized Graph Clustering via Path-based Similarity and Degree Descent Criterion
Dec 07, 2023
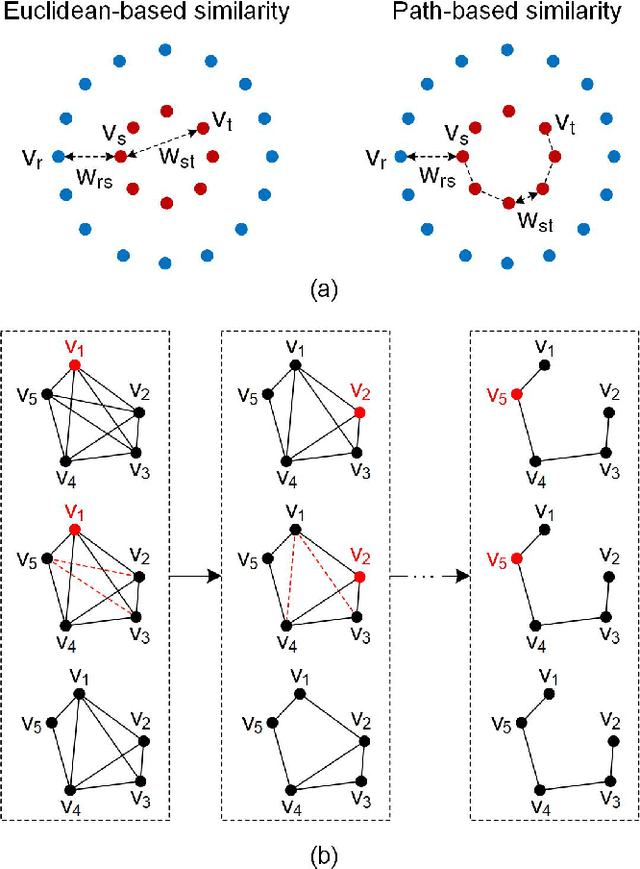
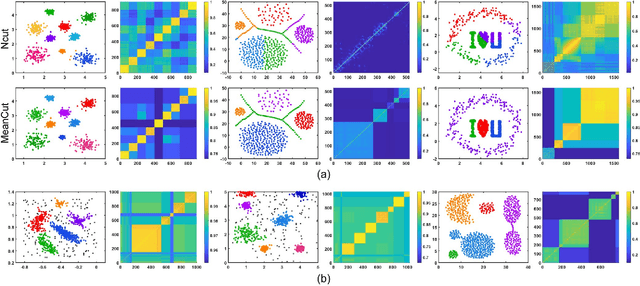
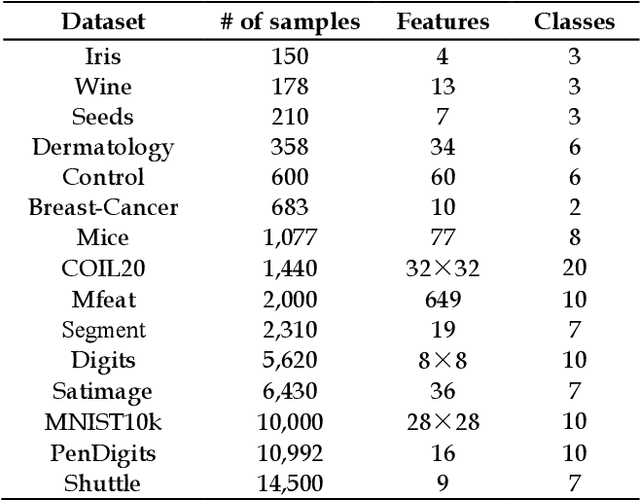
Abstract:As the most typical graph clustering method, spectral clustering is popular and attractive due to the remarkable performance, easy implementation, and strong adaptability. Classical spectral clustering measures the edge weights of graph using pairwise Euclidean-based metric, and solves the optimal graph partition by relaxing the constraints of indicator matrix and performing Laplacian decomposition. However, Euclidean-based similarity might cause skew graph cuts when handling non-spherical data distributions, and the relaxation strategy introduces information loss. Meanwhile, spectral clustering requires specifying the number of clusters, which is hard to determine without enough prior knowledge. In this work, we leverage the path-based similarity to enhance intra-cluster associations, and propose MeanCut as the objective function and greedily optimize it in degree descending order for a nondestructive graph partition. This algorithm enables the identification of arbitrary shaped clusters and is robust to noise. To reduce the computational complexity of similarity calculation, we transform optimal path search into generating the maximum spanning tree (MST), and develop a fast MST (FastMST) algorithm to further improve its time-efficiency. Moreover, we define a density gradient factor (DGF) for separating the weakly connected clusters. The validity of our algorithm is demonstrated by testifying on real-world benchmarks and application of face recognition. The source code of MeanCut is available at https://github.com/ZPGuiGroupWhu/MeanCut-Clustering.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge