David A Knowles
Active Learning in CNNs via Expected Improvement Maximization
Nov 27, 2020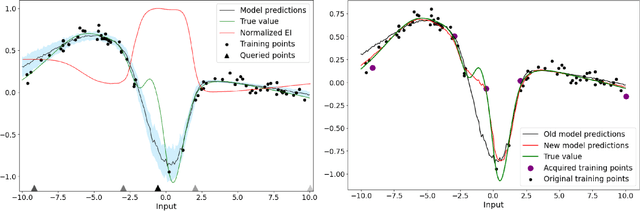

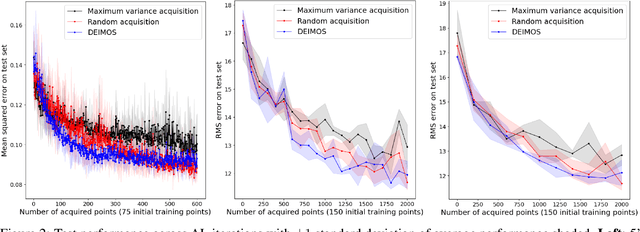
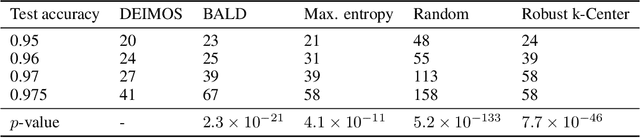
Abstract:Deep learning models such as Convolutional Neural Networks (CNNs) have demonstrated high levels of effectiveness in a variety of domains, including computer vision and more recently, computational biology. However, training effective models often requires assembling and/or labeling large datasets, which may be prohibitively time-consuming or costly. Pool-based active learning techniques have the potential to mitigate these issues, leveraging models trained on limited data to selectively query unlabeled data points from a pool in an attempt to expedite the learning process. Here we present "Dropout-based Expected IMprOvementS" (DEIMOS), a flexible and computationally-efficient approach to active learning that queries points that are expected to maximize the model's improvement across a representative sample of points. The proposed framework enables us to maintain a prediction covariance matrix capturing model uncertainty, and to dynamically update this matrix in order to generate diverse batches of points in the batch-mode setting. Our active learning results demonstrate that DEIMOS outperforms several existing baselines across multiple regression and classification tasks taken from computer vision and genomics.
A New Distribution on the Simplex with Auto-Encoding Applications
May 28, 2019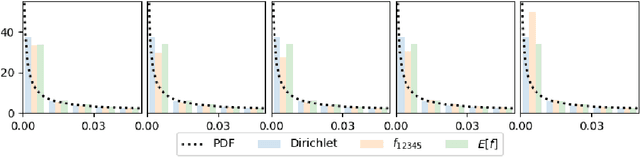
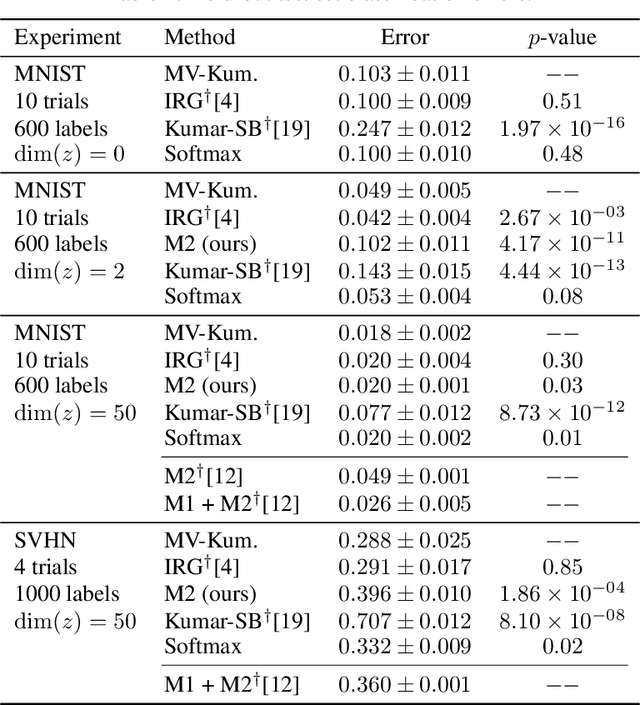
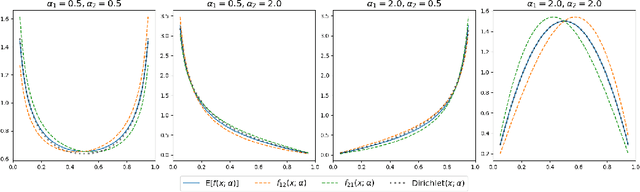
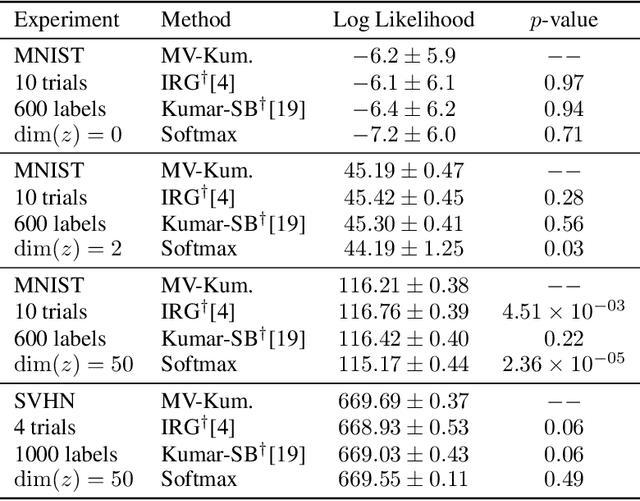
Abstract:We construct a new distribution for the simplex using the Kumaraswamy distribution and an ordered stick-breaking process. We explore and develop the theoretical properties of this new distribution and prove that it exhibits symmetry under the same conditions as the well-known Dirichlet. Like the Dirichlet, the new distribution is adept at capturing sparsity but, unlike the Dirichlet, has an exact and closed form reparameterization--making it well suited for deep variational Bayesian modeling. We demonstrate the distribution's utility in a variety of semi-supervised auto-encoding tasks. In all cases, the resulting models achieve competitive performance commensurate with their simplicity, use of explicit probability models, and abstinence from adversarial training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge