Danny Eitan
Enhancing Causal Estimation through Unlabeled Offline Data
Feb 16, 2022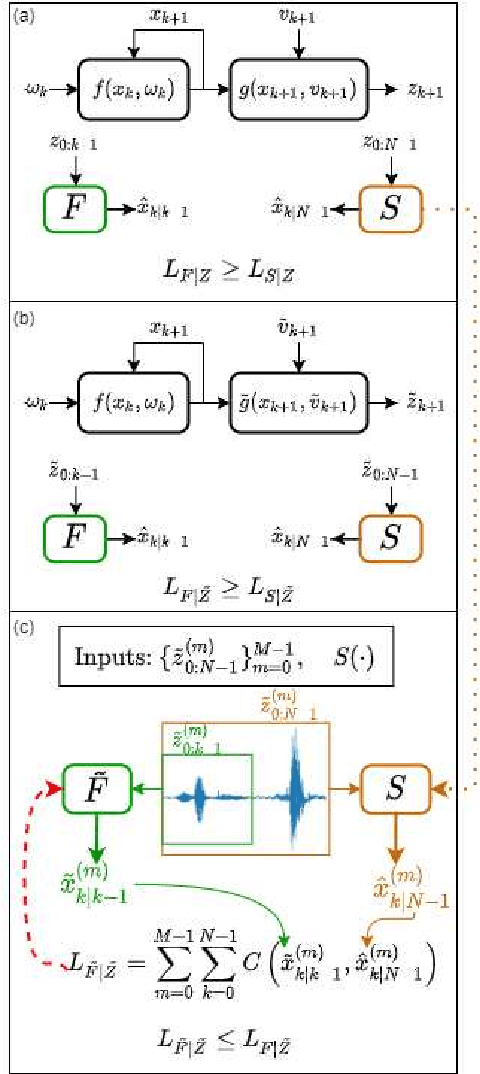
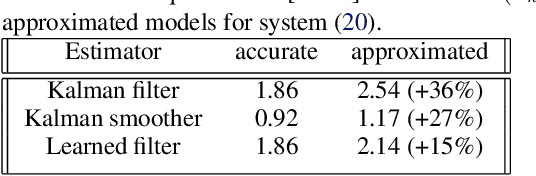
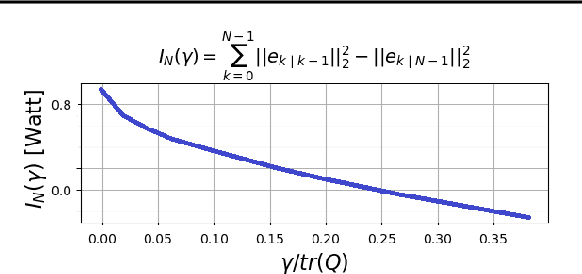
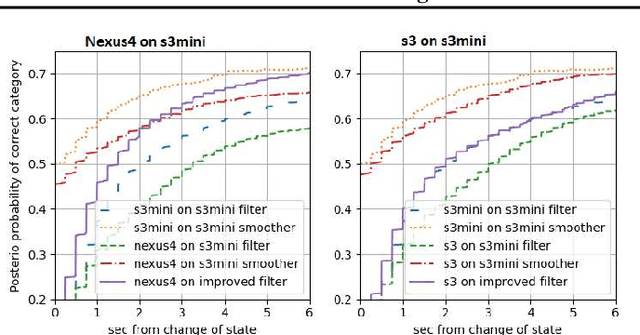
Abstract:Consider a situation where a new patient arrives in the Intensive Care Unit (ICU) and is monitored by multiple sensors. We wish to assess relevant unmeasured physiological variables (e.g., cardiac contractility and output and vascular resistance) that have a strong effect on the patients diagnosis and treatment. We do not have any information about this specific patient, but, extensive offline information is available about previous patients, that may only be partially related to the present patient (a case of dataset shift). This information constitutes our prior knowledge, and is both partial and approximate. The basic question is how to best use this prior knowledge, combined with online patient data, to assist in diagnosing the current patient most effectively. Our proposed approach consists of three stages: (i) Use the abundant offline data in order to create both a non-causal and a causal estimator for the relevant unmeasured physiological variables. (ii) Based on the non-causal estimator constructed, and a set of measurements from a new group of patients, we construct a causal filter that provides higher accuracy in the prediction of the hidden physiological variables for this new set of patients. (iii) For any new patient arriving in the ICU, we use the constructed filter in order to predict relevant internal variables. Overall, this strategy allows us to make use of the abundantly available offline data in order to enhance causal estimation for newly arriving patients. We demonstrate the effectiveness of this methodology on a (non-medical) real-world task, in situations where the offline data is only partially related to the new observations. We provide a mathematical analysis of the merits of the approach in a linear setting of Kalman filtering and smoothing, demonstrating its utility.
Continuous Forecasting via Neural Eigen Decomposition of Stochastic Dynamics
Feb 02, 2022



Abstract:Motivated by a real-world problem of blood coagulation control in Heparin-treated patients, we use Stochastic Differential Equations (SDEs) to formulate a new class of sequential prediction problems -- with an unknown latent space, unknown non-linear dynamics, and irregular sparse observations. We introduce the Neural Eigen-SDE (NESDE) algorithm for sequential prediction with sparse observations and adaptive dynamics. NESDE applies eigen-decomposition to the dynamics model to allow efficient frequent predictions given sparse observations. In addition, NESDE uses a learning mechanism for adaptive dynamics model, which handles changes in the dynamics both between sequences and within sequences. We demonstrate the accuracy and efficacy of NESDE for both synthetic problems and real-world data. In particular, to the best of our knowledge, we are the first to provide a patient-adapted prediction for blood coagulation following Heparin dosing in the MIMIC-IV dataset. Finally, we publish a simulated gym environment based on our prediction model, for experimentation in algorithms for blood coagulation control.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge