Daniel Israel
Enabling Autoregressive Models to Fill In Masked Tokens
Feb 09, 2025Abstract:Historically, LLMs have been trained using either autoregressive (AR) or masked language modeling (MLM) objectives, with AR models gaining dominance in recent years. However, AR models are inherently incapable of masked infilling, which is the ability to predict masked tokens between past and future context. In contrast, MLM models suffer from intrinsic computational inefficiencies during both training and inference that hinder their scalability. This work introduces MARIA (Masked and Autoregressive Infilling Architecture), a novel approach that leverages the strengths of both paradigms to achieve state-of-the-art masked infilling performance. MARIA combines a pre-trained MLM and AR model by training a linear decoder that takes their concatenated hidden states as input. This minimal modification enables the AR model to perform infilling while retaining its inherent advantages in terms of faster inference with KV caching. Our results demonstrate that MARIA significantly outperforms existing methods, namely discrete diffusion models, on masked infilling tasks.
MedMax: Mixed-Modal Instruction Tuning for Training Biomedical Assistants
Dec 17, 2024Abstract:Recent advancements in mixed-modal generative models have enabled flexible integration of information across image-text content. These models have opened new avenues for developing unified biomedical assistants capable of analyzing biomedical images, answering complex questions about them, and predicting the impact of medical procedures on a patient's health. However, existing resources face challenges such as limited data availability, narrow domain coverage, and restricted sources (e.g., medical papers). To address these gaps, we present MedMax, the first large-scale multimodal biomedical instruction-tuning dataset for mixed-modal foundation models. With 1.47 million instances, MedMax encompasses a diverse range of tasks, including multimodal content generation (interleaved image-text data), biomedical image captioning and generation, visual chatting, and report understanding. These tasks span diverse medical domains such as radiology and histopathology. Subsequently, we fine-tune a mixed-modal foundation model on the MedMax dataset, achieving significant performance improvements: a 26% gain over the Chameleon model and an 18.3% improvement over GPT-4o across 12 downstream biomedical visual question-answering tasks. Additionally, we introduce a unified evaluation suite for biomedical tasks, providing a robust framework to guide the development of next-generation mixed-modal biomedical AI assistants.
Prepacking: A Simple Method for Fast Prefilling and Increased Throughput in Large Language Models
Apr 15, 2024



Abstract:During inference for transformer-based large language models (LLM), prefilling is the computation of the key-value (KV) cache for input tokens in the prompt prior to autoregressive generation. For longer input prompt lengths, prefilling will incur a significant overhead on decoding time. In this work, we highlight the following pitfall of prefilling: for batches containing high-varying prompt lengths, significant computation is wasted by the standard practice of padding sequences to the maximum length. As LLMs increasingly support longer context lengths, potentially up to 10 million tokens, variations in prompt lengths within a batch become more pronounced. To address this, we propose Prepacking, a simple yet effective method to optimize prefilling computation. To avoid redundant computation on pad tokens, prepacking combines prompts of varying lengths into a sequence and packs multiple sequences into a compact batch using a bin-packing algorithm. It then modifies the attention mask and positional encoding to compute multiple prefilled KV-caches for multiple prompts within a single sequence. On standard curated dataset containing prompts with varying lengths, we obtain a significant speed and memory efficiency improvements as compared to the default padding-based prefilling computation within Huggingface across a range of base model configurations and inference serving scenarios.
High Dimensional Causal Inference with Variational Backdoor Adjustment
Oct 09, 2023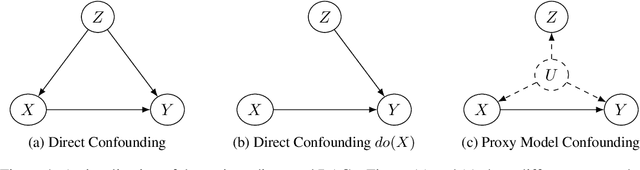
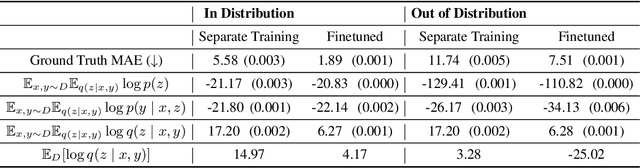
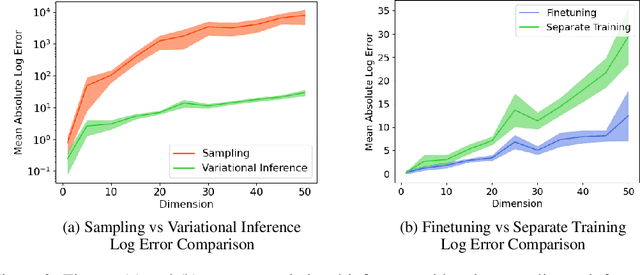
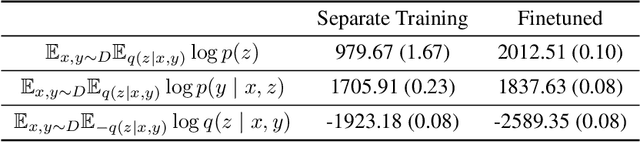
Abstract:Backdoor adjustment is a technique in causal inference for estimating interventional quantities from purely observational data. For example, in medical settings, backdoor adjustment can be used to control for confounding and estimate the effectiveness of a treatment. However, high dimensional treatments and confounders pose a series of potential pitfalls: tractability, identifiability, optimization. In this work, we take a generative modeling approach to backdoor adjustment for high dimensional treatments and confounders. We cast backdoor adjustment as an optimization problem in variational inference without reliance on proxy variables and hidden confounders. Empirically, our method is able to estimate interventional likelihood in a variety of high dimensional settings, including semi-synthetic X-ray medical data. To the best of our knowledge, this is the first application of backdoor adjustment in which all the relevant variables are high dimensional.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge