Daniel Catchpoole
Inferring Actual Treatment Pathways from Patient Records
Sep 05, 2023



Abstract:Treatment pathways are step-by-step plans outlining the recommended medical care for specific diseases; they get revised when different treatments are found to improve patient outcomes. Examining health records is an important part of this revision process, but inferring patients' actual treatments from health data is challenging due to complex event-coding schemes and the absence of pathway-related annotations. This study aims to infer the actual treatment steps for a particular patient group from administrative health records (AHR) - a common form of tabular healthcare data - and address several technique- and methodology-based gaps in treatment pathway-inference research. We introduce Defrag, a method for examining AHRs to infer the real-world treatment steps for a particular patient group. Defrag learns the semantic and temporal meaning of healthcare event sequences, allowing it to reliably infer treatment steps from complex healthcare data. To our knowledge, Defrag is the first pathway-inference method to utilise a neural network (NN), an approach made possible by a novel, self-supervised learning objective. We also developed a testing and validation framework for pathway inference, which we use to characterise and evaluate Defrag's pathway inference ability and compare against baselines. We demonstrate Defrag's effectiveness by identifying best-practice pathway fragments for breast cancer, lung cancer, and melanoma in public healthcare records. Additionally, we use synthetic data experiments to demonstrate the characteristics of the Defrag method, and to compare Defrag to several baselines where it significantly outperforms non-NN-based methods. Defrag significantly outperforms several existing pathway-inference methods and offers an innovative and effective approach for inferring treatment pathways from AHRs. Open-source code is provided to encourage further research in this area.
Machine Learning for Administrative Health Records: A Systematic Review of Techniques and Applications
Aug 27, 2023



Abstract:Machine learning provides many powerful and effective techniques for analysing heterogeneous electronic health records (EHR). Administrative Health Records (AHR) are a subset of EHR collected for administrative purposes, and the use of machine learning on AHRs is a growing subfield of EHR analytics. Existing reviews of EHR analytics emphasise that the data-modality of the EHR limits the breadth of suitable machine learning techniques, and pursuable healthcare applications. Despite emphasising the importance of data modality, the literature fails to analyse which techniques and applications are relevant to AHRs. AHRs contain uniquely well-structured, categorically encoded records which are distinct from other data-modalities captured by EHRs, and they can provide valuable information pertaining to how patients interact with the healthcare system. This paper systematically reviews AHR-based research, analysing 70 relevant studies and spanning multiple databases. We identify and analyse which machine learning techniques are applied to AHRs and which health informatics applications are pursued in AHR-based research. We also analyse how these techniques are applied in pursuit of each application, and identify the limitations of these approaches. We find that while AHR-based studies are disconnected from each other, the use of AHRs in health informatics research is substantial and accelerating. Our synthesis of these studies highlights the utility of AHRs for pursuing increasingly complex and diverse research objectives despite a number of pervading data- and technique-based limitations. Finally, through our findings, we propose a set of future research directions that can enhance the utility of AHR data and machine learning techniques for health informatics research.
Beyond Topics: Discovering Latent Healthcare Objectives from Event Sequences
Oct 04, 2021

Abstract:A meaningful understanding of clinical protocols and patient pathways helps improve healthcare outcomes. Electronic health records (EHR) reflect real-world treatment behaviours that are used to enhance healthcare management but present challenges; protocols and pathways are often loosely defined and with elements frequently not recorded in EHRs, complicating the enhancement. To solve this challenge, healthcare objectives associated with healthcare management activities can be indirectly observed in EHRs as latent topics. Topic models, such as Latent Dirichlet Allocation (LDA), are used to identify latent patterns in EHR data. However, they do not examine the ordered nature of EHR sequences, nor do they appraise individual events in isolation. Our novel approach, the Categorical Sequence Encoder (CaSE) addresses these shortcomings. The sequential nature of EHRs is captured by CaSE's event-level representations, revealing latent healthcare objectives. In synthetic EHR sequences, CaSE outperforms LDA by up to 37% at identifying healthcare objectives. In the real-world MIMIC-III dataset, CaSE identifies meaningful representations that could critically enhance protocol and pathway development.
Relational Autoencoder for Feature Extraction
Feb 09, 2018
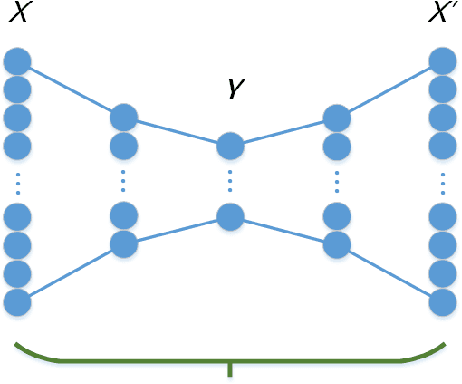
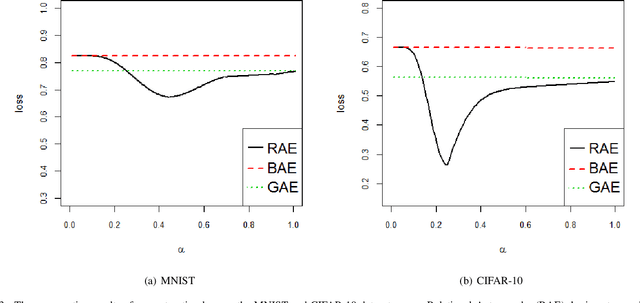
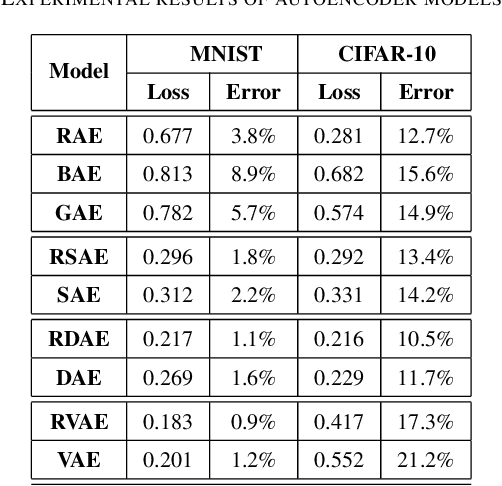
Abstract:Feature extraction becomes increasingly important as data grows high dimensional. Autoencoder as a neural network based feature extraction method achieves great success in generating abstract features of high dimensional data. However, it fails to consider the relationships of data samples which may affect experimental results of using original and new features. In this paper, we propose a Relation Autoencoder model considering both data features and their relationships. We also extend it to work with other major autoencoder models including Sparse Autoencoder, Denoising Autoencoder and Variational Autoencoder. The proposed relational autoencoder models are evaluated on a set of benchmark datasets and the experimental results show that considering data relationships can generate more robust features which achieve lower construction loss and then lower error rate in further classification compared to the other variants of autoencoders.
* IJCNN-2017
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge