Corey Wang
HARDMath: A Benchmark Dataset for Challenging Problems in Applied Mathematics
Oct 13, 2024



Abstract:Advanced applied mathematics problems are underrepresented in existing Large Language Model (LLM) benchmark datasets. To address this, we introduce HARDMath, a dataset inspired by a graduate course on asymptotic methods, featuring challenging applied mathematics problems that require analytical approximation techniques. These problems demand a combination of mathematical reasoning, computational tools, and subjective judgment, making them difficult for LLMs. Our framework auto-generates a large number of problems with solutions validated against numerical ground truths. We evaluate both open- and closed-source LLMs on HARDMath-mini, a sub-sampled test set of 366 problems, as well as on 40 word problems formulated in applied science contexts. Even leading closed-source models like GPT-4 achieve only 43.8% overall accuracy with few-shot Chain-of-Thought prompting, and all models demonstrate significantly lower performance compared to results on existing mathematics benchmark datasets. We additionally conduct a detailed error analysis to gain insights into the failure cases of LLMs. These results demonstrate limitations of current LLM performance on advanced graduate-level applied math problems and underscore the importance of datasets like HARDMath to advance mathematical abilities of LLMs.
Contrastive Image Synthesis and Self-supervised Feature Adaptation for Cross-Modality Biomedical Image Segmentation
Jul 27, 2022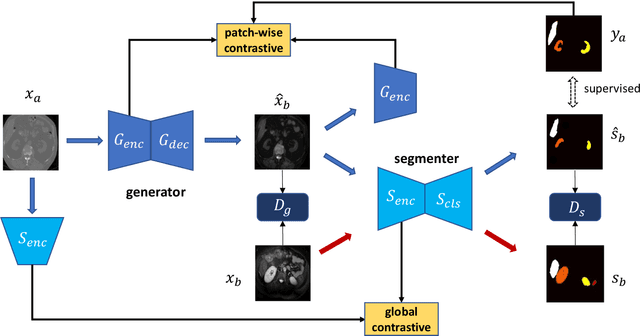



Abstract:This work presents a novel framework CISFA (Contrastive Image synthesis and Self-supervised Feature Adaptation)that builds on image domain translation and unsupervised feature adaptation for cross-modality biomedical image segmentation. Different from existing works, we use a one-sided generative model and add a weighted patch-wise contrastive loss between sampled patches of the input image and the corresponding synthetic image, which serves as shape constraints. Moreover, we notice that the generated images and input images share similar structural information but are in different modalities. As such, we enforce contrastive losses on the generated images and the input images to train the encoder of a segmentation model to minimize the discrepancy between paired images in the learned embedding space. Compared with existing works that rely on adversarial learning for feature adaptation, such a method enables the encoder to learn domain-independent features in a more explicit way. We extensively evaluate our methods on segmentation tasks containing CT and MRI images for abdominal cavities and whole hearts. Experimental results show that the proposed framework not only outputs synthetic images with less distortion of organ shapes, but also outperforms state-of-the-art domain adaptation methods by a large margin.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge