Colin Ophus
Unsupervised clustering algorithm for efficient processing of 4D-STEM and 5D-STEM data
Jan 24, 2026Abstract:Four-dimensional scanning transmission electron microscopy (4D-STEM) enables mapping of diffraction information with nanometer-scale spatial resolution, offering detailed insight into local structure, orientation, and strain. However, as data dimensionality and sampling density increase, particularly for in situ scanning diffraction experiments (5D-STEM), robust segmentation of spatially coherent regions becomes essential for efficient and physically meaningful analysis. Here, we introduce a clustering framework that identifies crystallographically distinct domains from 4D-STEM datasets. By using local diffraction-pattern similarity as a metric, the method extracts closed contours delineating regions of coherent structural behavior. This approach produces cluster-averaged diffraction patterns that improve signal-to-noise and reduce data volume by orders of magnitude, enabling rapid and accurate orientation, phase, and strain mapping. We demonstrate the applicability of this approach to in situ liquid-cell 4D-STEM data of gold nanoparticle growth. Our method provides a scalable and generalizable route for spatially coherent segmentation, data compression, and quantitative structure-strain mapping across diverse 4D-STEM modalities. The full analysis code and example workflows are publicly available to support reproducibility and reuse.
A Gaussian Parameterization for Direct Atomic Structure Identification in Electron Tomography
Dec 17, 2025Abstract:Atomic electron tomography (AET) enables the determination of 3D atomic structures by acquiring a sequence of 2D tomographic projection measurements of a particle and then computationally solving for its underlying 3D representation. Classical tomography algorithms solve for an intermediate volumetric representation that is post-processed into the atomic structure of interest. In this paper, we reformulate the tomographic inverse problem to solve directly for the locations and properties of individual atoms. We parameterize an atomic structure as a collection of Gaussians, whose positions and properties are learnable. This representation imparts a strong physical prior on the learned structure, which we show yields improved robustness to real-world imaging artifacts. Simulated experiments and a proof-of-concept result on experimentally-acquired data confirm our method's potential for practical applications in materials characterization and analysis with Transmission Electron Microscopy (TEM). Our code is available at https://github.com/nalinimsingh/gaussian-atoms.
Missing Wedge Inpainting and Joint Alignment in Electron Tomography through Implicit Neural Representations
Dec 08, 2025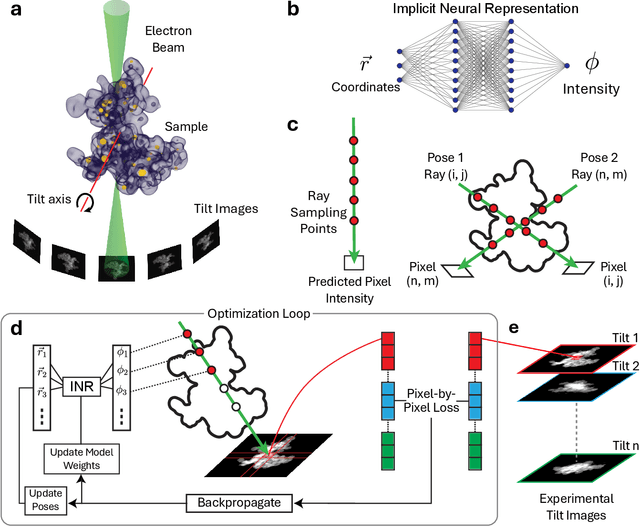
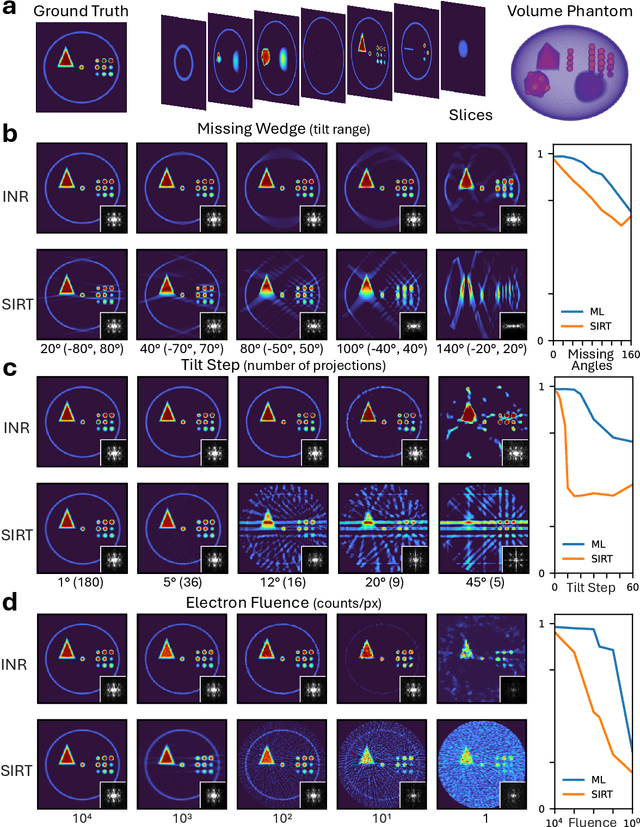
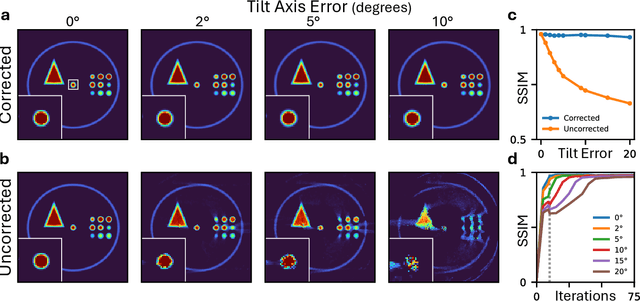
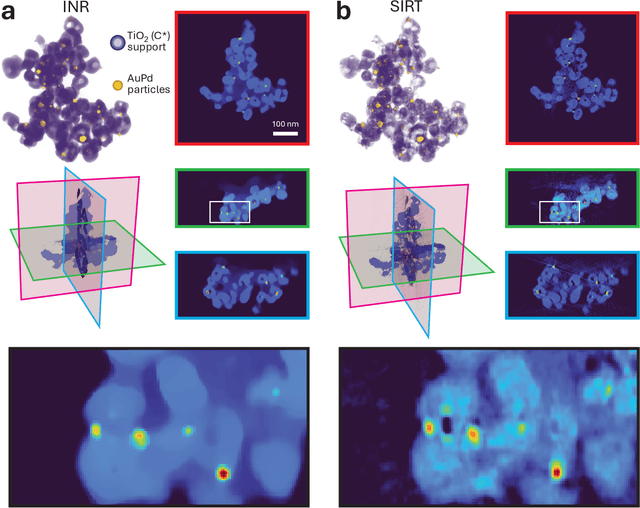
Abstract:Electron tomography is a powerful tool for understanding the morphology of materials in three dimensions, but conventional reconstruction algorithms typically suffer from missing-wedge artifacts and data misalignment imposed by experimental constraints. Recently proposed supervised machine-learning-enabled reconstruction methods to address these challenges rely on training data and are therefore difficult to generalize across materials systems. We propose a fully self-supervised implicit neural representation (INR) approach using a neural network as a regularizer. Our approach enables fast inline alignment through pose optimization, missing wedge inpainting, and denoising of low dose datasets via model regularization using only a single dataset. We apply our method to simulated and experimental data and show that it produces high-quality tomograms from diverse and information limited datasets. Our results show that INR-based self-supervised reconstructions offer high fidelity reconstructions with minimal user input and preprocessing, and can be readily applied to a wide variety of materials samples and experimental parameters.
Deep generative priors for robust and efficient electron ptychography
Nov 11, 2025Abstract:Electron ptychography enables dose-efficient atomic-resolution imaging, but conventional reconstruction algorithms suffer from noise sensitivity, slow convergence, and extensive manual hyperparameter tuning for regularization, especially in three-dimensional multislice reconstructions. We introduce a deep generative prior (DGP) framework for electron ptychography that uses the implicit regularization of convolutional neural networks to address these challenges. Two DGPs parameterize the complex-valued sample and probe within an automatic-differentiation mixed-state multislice forward model. Compared to pixel-based reconstructions, DGPs offer four key advantages: (i) greater noise robustness and improved information limits at low dose; (ii) markedly faster convergence, especially at low spatial frequencies; (iii) improved depth regularization; and (iv) minimal user-specified regularization. The DGP framework promotes spatial coherence and suppresses high-frequency noise without extensive tuning, and a pre-training strategy stabilizes reconstructions. Our results establish DGP-enabled ptychography as a robust approach that reduces expertise barriers and computational cost, delivering robust, high-resolution imaging across diverse materials and biological systems.
Disentangling multiple scattering with deep learning: application to strain mapping from electron diffraction patterns
Feb 01, 2022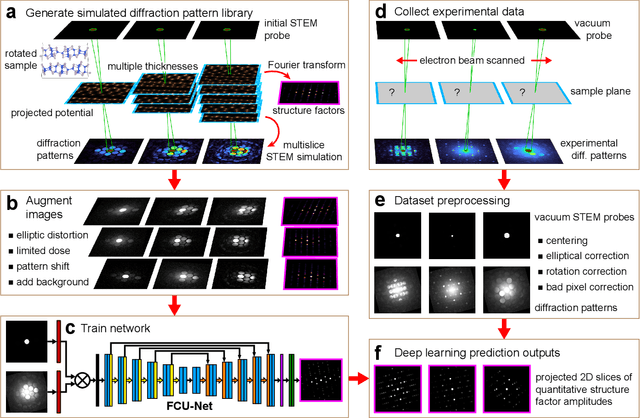
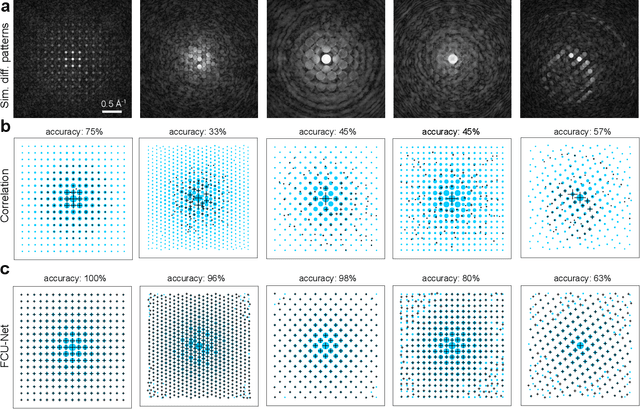
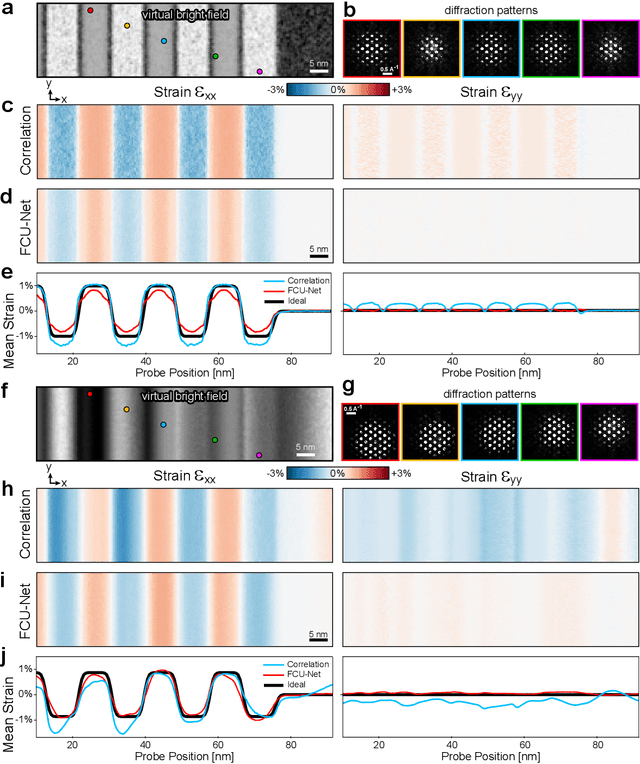

Abstract:Implementation of a fast, robust, and fully-automated pipeline for crystal structure determination and underlying strain mapping for crystalline materials is important for many technological applications. Scanning electron nanodiffraction offers a procedure for identifying and collecting strain maps with good accuracy and high spatial resolutions. However, the application of this technique is limited, particularly in thick samples where the electron beam can undergo multiple scattering, which introduces signal nonlinearities. Deep learning methods have the potential to invert these complex signals, but previous implementations are often trained only on specific crystal systems or a small subset of the crystal structure and microscope parameter phase space. In this study, we implement a Fourier space, complex-valued deep neural network called FCU-Net, to invert highly nonlinear electron diffraction patterns into the corresponding quantitative structure factor images. We trained the FCU-Net using over 200,000 unique simulated dynamical diffraction patterns which include many different combinations of crystal structures, orientations, thicknesses, microscope parameters, and common experimental artifacts. We evaluated the trained FCU-Net model against simulated and experimental 4D-STEM diffraction datasets, where it substantially out-performs conventional analysis methods. Our simulated diffraction pattern library, implementation of FCU-Net, and trained model weights are freely available in open source repositories, and can be adapted to many different diffraction measurement problems.
Distributed Reconstruction Algorithm for Electron Tomography with Multiple-scattering Samples
Oct 15, 2021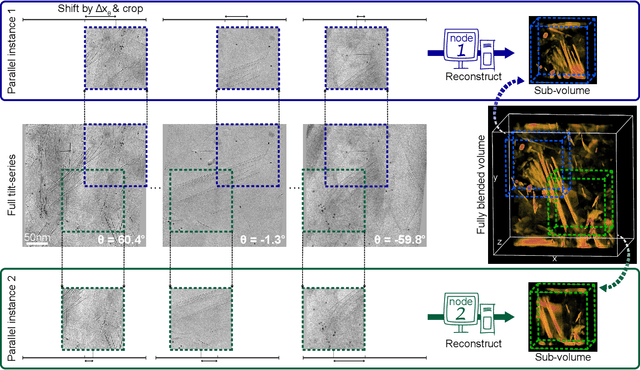
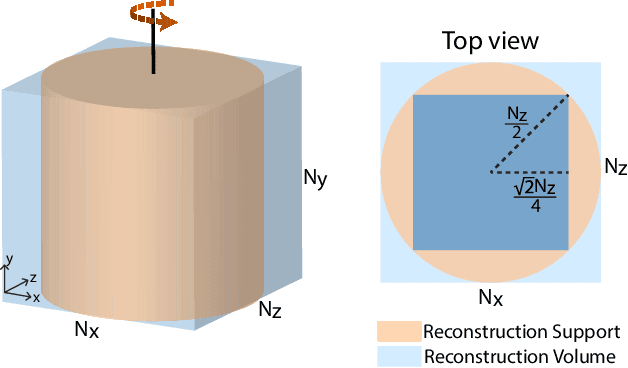
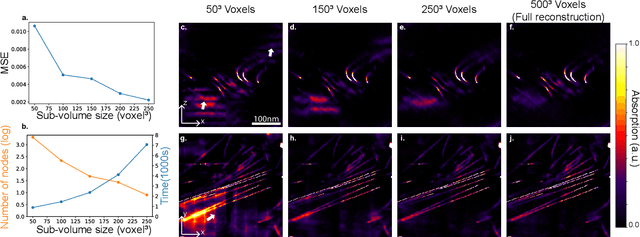

Abstract:Three-dimensional electron tomography is used to understand the structure and properties of samples in chemistry, materials science, geoscience, and biology. With the recent development of high-resolution detectors and algorithms that can account for multiple-scattering events, thicker samples can be examined at finer resolution, resulting in larger reconstruction volumes than previously possible. In this work, we propose a distributed computing framework that reconstructs large volumes by decomposing a projected tilt-series into smaller datasets such that sub-volumes can be simultaneously reconstructed on separate compute nodes using a cluster. We demonstrate our method by reconstructing a multiple-scattering layered clay (montmorillonite) sample at high resolution from a large field-of-view tilt-series phase contrast transmission electron microscopty dataset.
Deep Learning Segmentation of Complex Features in Atomic-Resolution Phase Contrast Transmission Electron Microscopy Images
Dec 09, 2020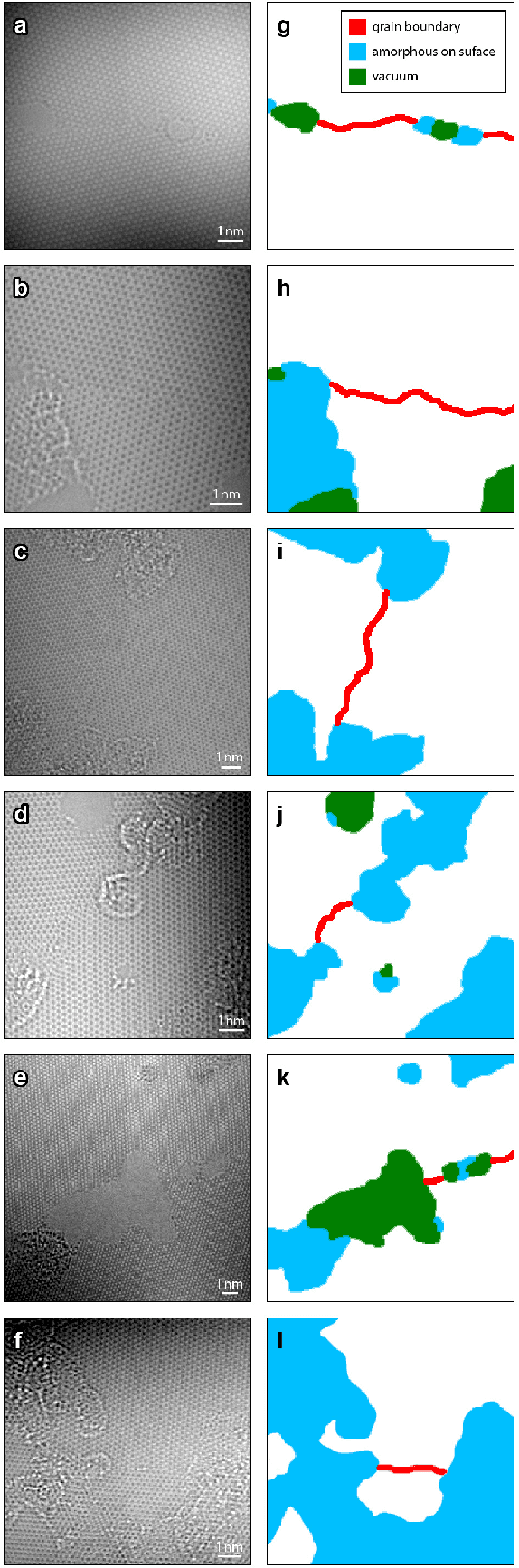
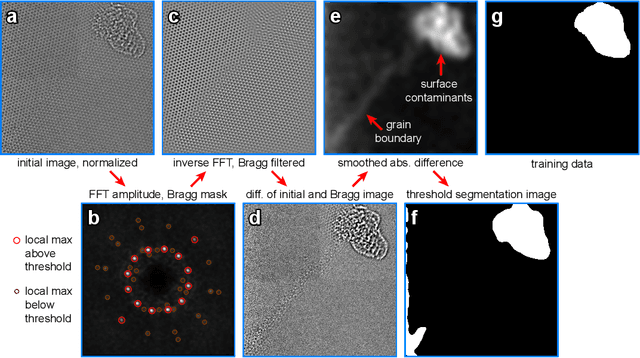
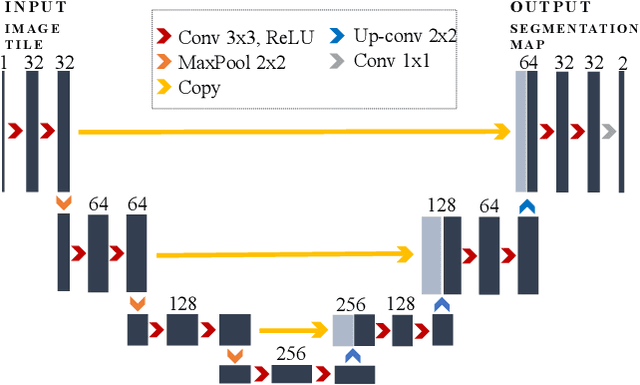
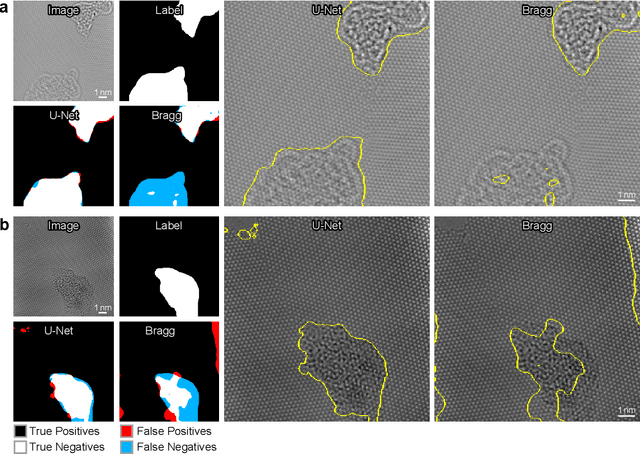
Abstract:Phase contrast transmission electron microscopy (TEM) is a powerful tool for imaging the local atomic structure of materials. TEM has been used heavily in studies of defect structures of 2D materials such as monolayer graphene due to its high dose efficiency. However, phase contrast imaging can produce complex nonlinear contrast, even for weakly-scattering samples. It is therefore difficult to develop fully-automated analysis routines for phase contrast TEM studies using conventional image processing tools. For automated analysis of large sample regions of graphene, one of the key problems is segmentation between the structure of interest and unwanted structures such as surface contaminant layers. In this study, we compare the performance of a conventional Bragg filtering method to a deep learning routine based on the U-Net architecture. We show that the deep learning method is more general, simpler to apply in practice, and produces more accurate and robust results than the conventional algorithm. We provide easily-adaptable source code for all results in this paper, and discuss potential applications for deep learning in fully-automated TEM image analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge