Claire Morgand
ARS IDF
Causal thinking for decision making on Electronic Health Records: why and how
Aug 03, 2023



Abstract:Accurate predictions, as with machine learning, may not suffice to provide optimal healthcare for every patient. Indeed, prediction can be driven by shortcuts in the data, such as racial biases. Causal thinking is needed for data-driven decisions. Here, we give an introduction to the key elements, focusing on routinely-collected data, electronic health records (EHRs) and claims data. Using such data to assess the value of an intervention requires care: temporal dependencies and existing practices easily confound the causal effect. We present a step-by-step framework to help build valid decision making from real-life patient records by emulating a randomized trial before individualizing decisions, eg with machine learning. Our framework highlights the most important pitfalls and considerations in analysing EHRs or claims data to draw causal conclusions. We illustrate the various choices in studying the effect of albumin on sepsis mortality in the Medical Information Mart for Intensive Care database (MIMIC-IV). We study the impact of various choices at every step, from feature extraction to causal-estimator selection. In a tutorial spirit, the code and the data are openly available.
Neural translation and automated recognition of ICD10 medical entities from natural language
Mar 27, 2020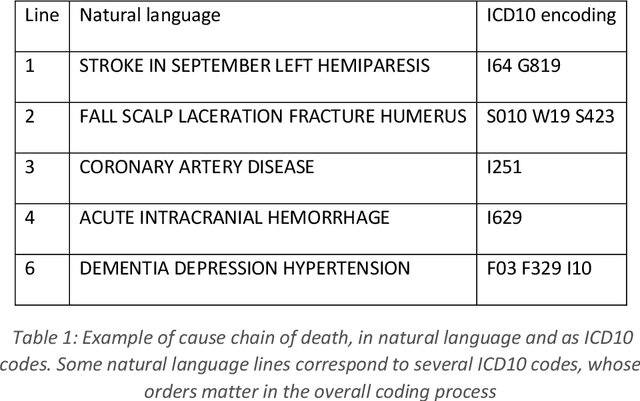
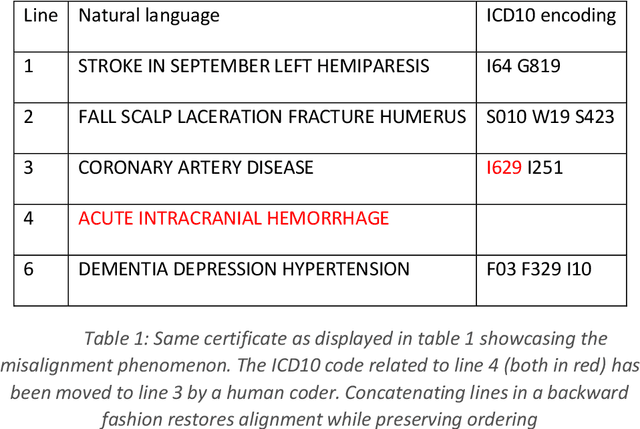
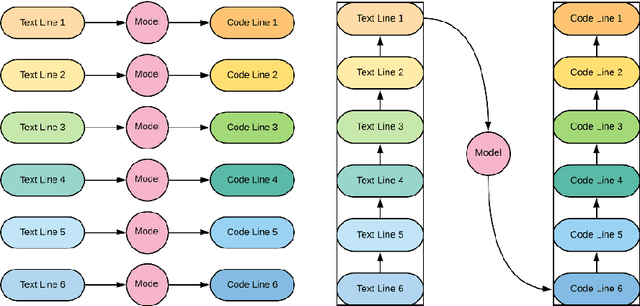

Abstract:The recognition of medical entities from natural language is an ubiquitous problem in the medical field, with applications ranging from medical act coding to the analysis of electronic health data for public health. It is however a complex task usually requiring human expert intervention, thus making it expansive and time consuming. The recent advances in artificial intelligence, specifically the raise of deep learning methods, has enabled computers to make efficient decisions on a number of complex problems, with the notable example of neural sequence models and their powerful applications in natural language processing. They however require a considerable amount of data to learn from, which is typically their main limiting factor. However, the C\'epiDc stores an exhaustive database of death certificates at the French national scale, amounting to several millions of natural language examples provided with their associated human coded medical entities available to the machine learning practitioner. This article investigates the applications of deep neural sequence models to the medical entity recognition from natural language problem.
A deep artificial neural network based model for underlying cause of death prediction from death certificates
Aug 26, 2019



Abstract:Underlying cause of death coding from death certificates is a process that is nowadays undertaken mostly by humans with a potential assistance from expert systems such as the Iris software. It is as a consequence an expensive process that can in addition suffer from geospatial discrepancies, thus severely impairing the comparability of death statistics at the international level. The recent advances in artificial intelligence, specifically the raise of deep learning methods, has enabled computers to make efficient decisions on a number of complex problem that were typically considered as out of reach without human assistance. They however require a considerable amount of data to learn from, which is typically their main limiting factor. However, the C\'epiDc stores an exhaustive database of death certificate at the French national scale, amounting to several millions training example available for the machine learning practitioner. This article presents a deep learning based tool for automated coding of the underlying cause of death from the data contained in death certificates with 97.8% accuracy, a substantial achievement compared to the Iris software and its 75% accuracy assessed on the same test examples. Such an improvement opens a whole field of new applications, from nosologist-level batch automated coding to international and temporal harmonization of cause of death statistics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge