Chun Meng
Research on Cervical Cancer p16/Ki-67 Immunohistochemical Dual-Staining Image Recognition Algorithm Based on YOLO
Dec 02, 2024



Abstract:The p16/Ki-67 dual staining method is a new approach for cervical cancer screening with high sensitivity and specificity. However, there are issues of mis-detection and inaccurate recognition when the YOLOv5s algorithm is directly applied to dual-stained cell images. This paper Proposes a novel cervical cancer dual-stained image recognition (DSIR-YOLO) model based on an YOLOv5. By fusing the Swin-Transformer module, GAM attention mechanism, multi-scale feature fusion, and EIoU loss function, the detection performance is significantly improved, with mAP@0.5 and mAP@0.5:0.95 reaching 92.6% and 70.5%, respectively. Compared with YOLOv5s in five-fold cross-validation, the accuracy, recall, mAP@0.5, and mAP@0.5:0.95 of the improved algorithm are increased by 2.3%, 4.1%, 4.3%, and 8.0%, respectively, with smaller variances and higher stability. Compared with other detection algorithms, DSIR-YOLO in this paper sacrifices some performance requirements to improve the network recognition effect. In addition, the influence of dataset quality on the detection results is studied. By controlling the sealing property of pixels, scale difference, unlabelled cells, and diagonal annotation, the model detection accuracy, recall, mAP@0.5, and mAP@0.5:0.95 are improved by 13.3%, 15.3%, 18.3%, and 30.5%, respectively.
Brain-wise Tumor Segmentation and Patient Overall Survival Prediction
Sep 15, 2019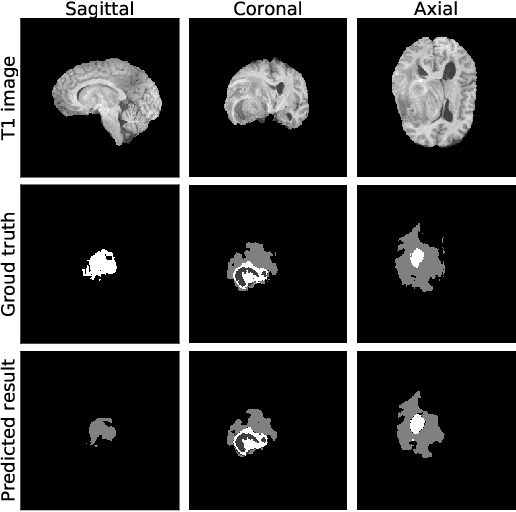
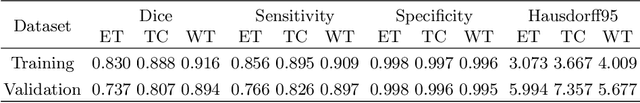
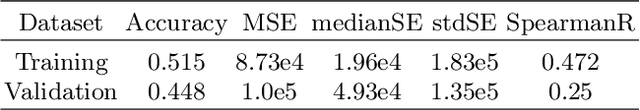
Abstract:Past few years have witnessed the prevalence of deep learning in many application scenarios, among which is medical image processing. Diagnosis and treatment of brain tumors require a delicate segmentation of brain tumors as a prerequisite. However, such kind of work conventionally costs cerebral surgeons a lot of precious time. Computer vision techniques could provide surgeons a relief from the tedious marking procedure. In this paper, a 3D U-net based deep learning model has been trained with the help of brain-wise normalization and patching strategies for the brain tumor segmentation task in BraTS 2019 competition. Dice coefficients for enhancing tumor, tumor core, and the whole tumor are 0.737, 0.807 and 0.894 respectively on validation dataset. Furthermore, numerical features extracted from predicted tumor labels have been used for the overall survival days prediction task. The prediction accuracy on validation dataset is 0.448.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge