Christian N. Kruse
DG-TTA: Out-of-domain medical image segmentation through Domain Generalization and Test-Time Adaptation
Dec 22, 2023



Abstract:Applying pre-trained medical segmentation models on out-of-domain images often yields predictions of insufficient quality. Several strategies have been proposed to maintain model performance, such as finetuning or unsupervised- and source-free domain adaptation. These strategies set restrictive requirements for data availability. In this study, we propose to combine domain generalization and test-time adaptation to create a highly effective approach for reusing pre-trained models in unseen target domains. Domain-generalized pre-training on source data is used to obtain the best initial performance in the target domain. We introduce the MIND descriptor previously used in image registration tasks as a further technique to achieve generalization and present superior performance for small-scale datasets compared to existing approaches. At test-time, high-quality segmentation for every single unseen scan is ensured by optimizing the model weights for consistency given different image augmentations. That way, our method enables separate use of source and target data and thus removes current data availability barriers. Moreover, the presented method is highly modular as it does not require specific model architectures or prior knowledge of involved domains and labels. We demonstrate this by integrating it into the nnUNet, which is currently the most popular and accurate framework for medical image segmentation. We employ multiple datasets covering abdominal, cardiac, and lumbar spine scans and compose several out-of-domain scenarios in this study. We demonstrate that our method, combined with pre-trained whole-body CT models, can effectively segment MR images with high accuracy in all of the aforementioned scenarios. Open-source code can be found here: https://github.com/multimodallearning/DG-TTA
CrossMoDA 2021 challenge: Benchmark of Cross-Modality Domain Adaptation techniques for Vestibular Schwnannoma and Cochlea Segmentation
Jan 08, 2022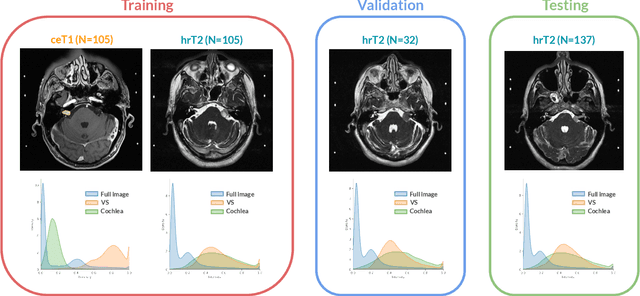
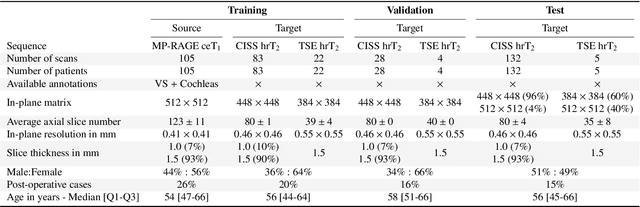
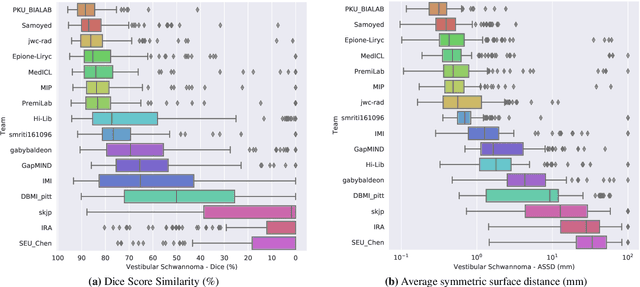
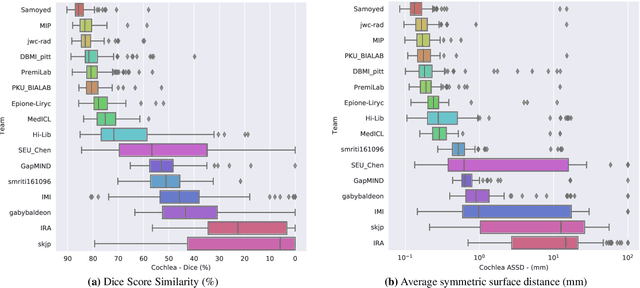
Abstract:Domain Adaptation (DA) has recently raised strong interests in the medical imaging community. While a large variety of DA techniques has been proposed for image segmentation, most of these techniques have been validated either on private datasets or on small publicly available datasets. Moreover, these datasets mostly addressed single-class problems. To tackle these limitations, the Cross-Modality Domain Adaptation (crossMoDA) challenge was organised in conjunction with the 24th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2021). CrossMoDA is the first large and multi-class benchmark for unsupervised cross-modality DA. The challenge's goal is to segment two key brain structures involved in the follow-up and treatment planning of vestibular schwannoma (VS): the VS and the cochleas. Currently, the diagnosis and surveillance in patients with VS are performed using contrast-enhanced T1 (ceT1) MRI. However, there is growing interest in using non-contrast sequences such as high-resolution T2 (hrT2) MRI. Therefore, we created an unsupervised cross-modality segmentation benchmark. The training set provides annotated ceT1 (N=105) and unpaired non-annotated hrT2 (N=105). The aim was to automatically perform unilateral VS and bilateral cochlea segmentation on hrT2 as provided in the testing set (N=137). A total of 16 teams submitted their algorithm for the evaluation phase. The level of performance reached by the top-performing teams is strikingly high (best median Dice - VS:88.4%; Cochleas:85.7%) and close to full supervision (median Dice - VS:92.5%; Cochleas:87.7%). All top-performing methods made use of an image-to-image translation approach to transform the source-domain images into pseudo-target-domain images. A segmentation network was then trained using these generated images and the manual annotations provided for the source image.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge