Cholmin Kang
Variability Matters : Evaluating inter-rater variability in histopathology for robust cell detection
Oct 11, 2022

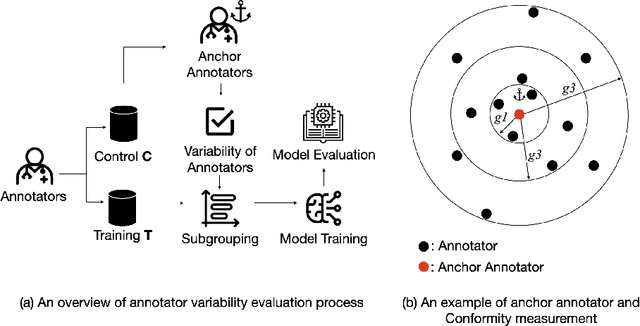
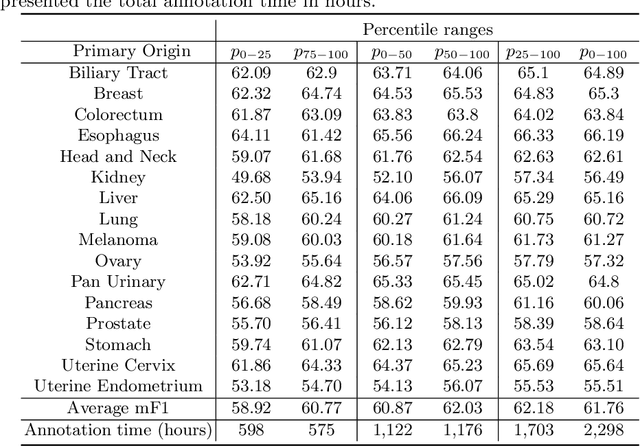
Abstract:Large annotated datasets have been a key component in the success of deep learning. However, annotating medical images is challenging as it requires expertise and a large budget. In particular, annotating different types of cells in histopathology suffer from high inter- and intra-rater variability due to the ambiguity of the task. Under this setting, the relation between annotators' variability and model performance has received little attention. We present a large-scale study on the variability of cell annotations among 120 board-certified pathologists and how it affects the performance of a deep learning model. We propose a method to measure such variability, and by excluding those annotators with low variability, we verify the trade-off between the amount of data and its quality. We found that naively increasing the data size at the expense of inter-rater variability does not necessarily lead to better-performing models in cell detection. Instead, decreasing the inter-rater variability with the expense of decreasing dataset size increased the model performance. Furthermore, models trained from data annotated with lower inter-labeler variability outperform those from higher inter-labeler variability. These findings suggest that the evaluation of the annotators may help tackle the fundamental budget issues in the histopathology domain
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge