Chi-Sing Ho
Interpretable Signal Analysis with Knockoffs Enhances Classification of Bacterial Raman Spectra
Jun 08, 2020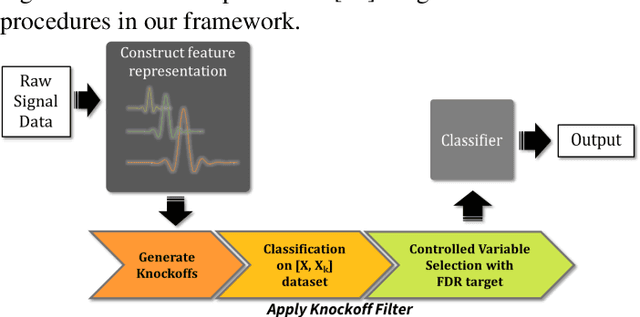
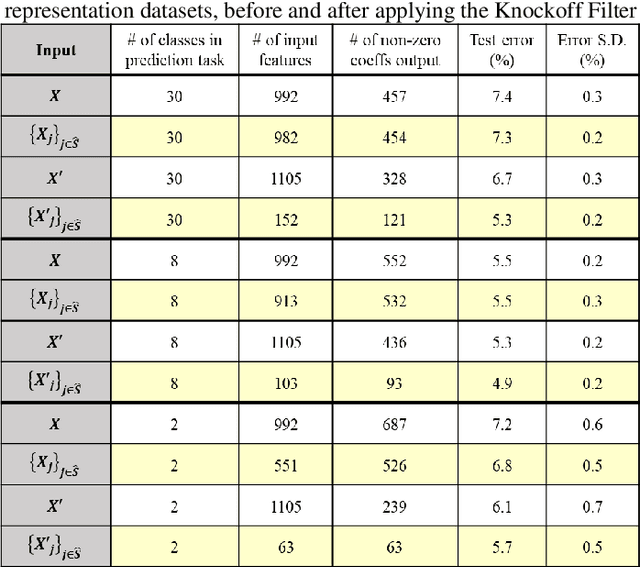

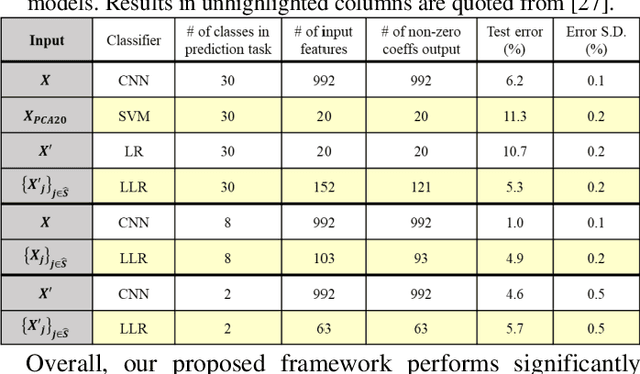
Abstract:Interpretability is important for many applications of machine learning to signal data, covering aspects such as how well a model fits the data, how accurately explanations are drawn from it, and how well these can be understood by people. Feature extraction and selection can improve model interpretability by identifying structures in the data that are both informative and intuitively meaningful. To this end, we propose a signal classification framework that combines feature extraction with feature selection using the knockoff filter, a method which provides guarantees on the false discovery rate (FDR) amongst selected features. We apply this to a dataset of Raman spectroscopy measurements from bacterial samples. Using a wavelet-based feature representation of the data and a logistic regression classifier, our framework achieves significantly higher predictive accuracy compared to using the original features as input. Benchmarking was also done with features obtained through principal components analysis, as well as the original features input into a neural network-based classifier. Our proposed framework achieved better predictive performance at the former task and comparable performance at the latter task, while offering the advantage of a more compact and human-interpretable set of features.
Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning
Jan 23, 2019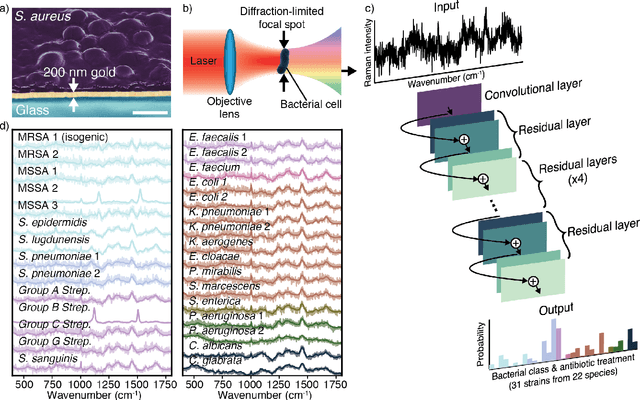


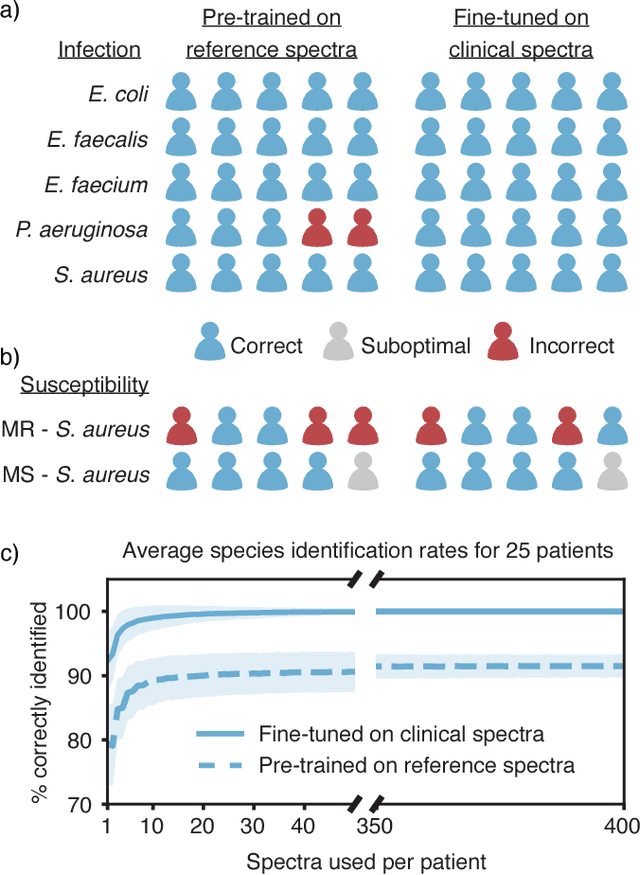
Abstract:Rapid identification of bacteria is essential to prevent the spread of infectious disease, help combat antimicrobial resistance, and improve patient outcomes. Raman optical spectroscopy promises to combine bacterial detection, identification, and antibiotic susceptibility testing in a single step. However, achieving clinically relevant speeds and accuracies remains challenging due to the weak Raman signal from bacterial cells and the large number of bacterial species and phenotypes. By amassing the largest known dataset of bacterial Raman spectra, we are able to apply state-of-the-art deep learning approaches to identify 30 of the most common bacterial pathogens from noisy Raman spectra, achieving antibiotic treatment identification accuracies of 99.0$\pm$0.1%. This novel approach distinguishes between methicillin-resistant and -susceptible isolates of Staphylococcus aureus (MRSA and MSSA) as well as a pair of isogenic MRSA and MSSA that are genetically identical apart from deletion of the mecA resistance gene, indicating the potential for culture-free detection of antibiotic resistance. Results from initial clinical validation are promising: using just 10 bacterial spectra from each of 25 isolates, we achieve 99.0$\pm$1.9% species identification accuracy. Our combined Raman-deep learning system represents an important proof-of-concept for rapid, culture-free identification of bacterial isolates and antibiotic resistance and could be readily extended for diagnostics on blood, urine, and sputum.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge