Chae Eun Lee
Voxel-wise Adversarial Semi-supervised Learning for Medical Image Segmentation
May 14, 2022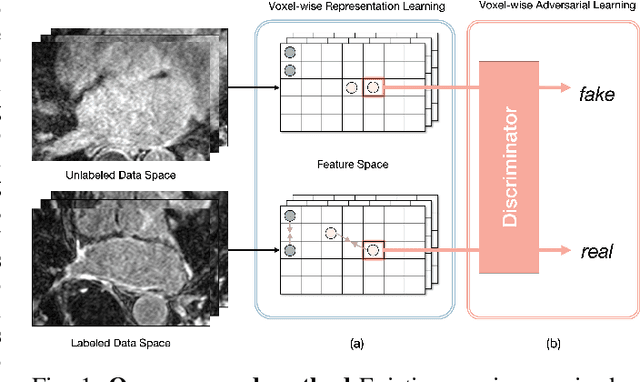
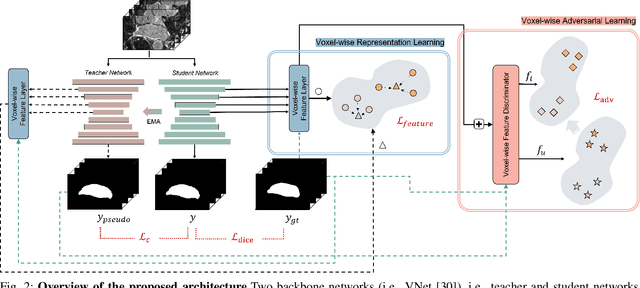
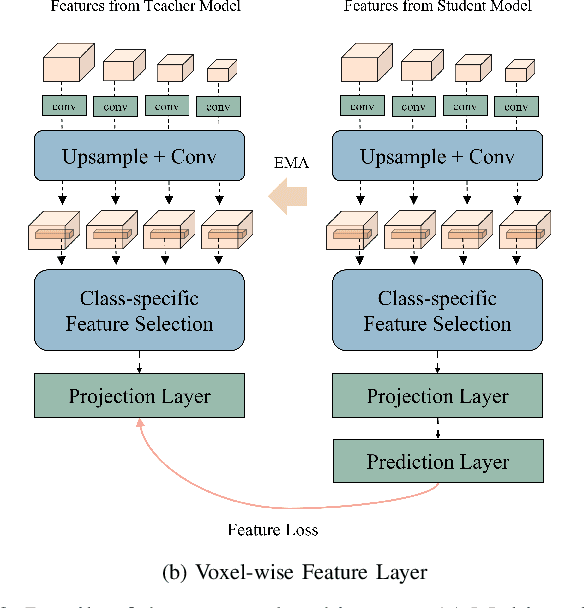
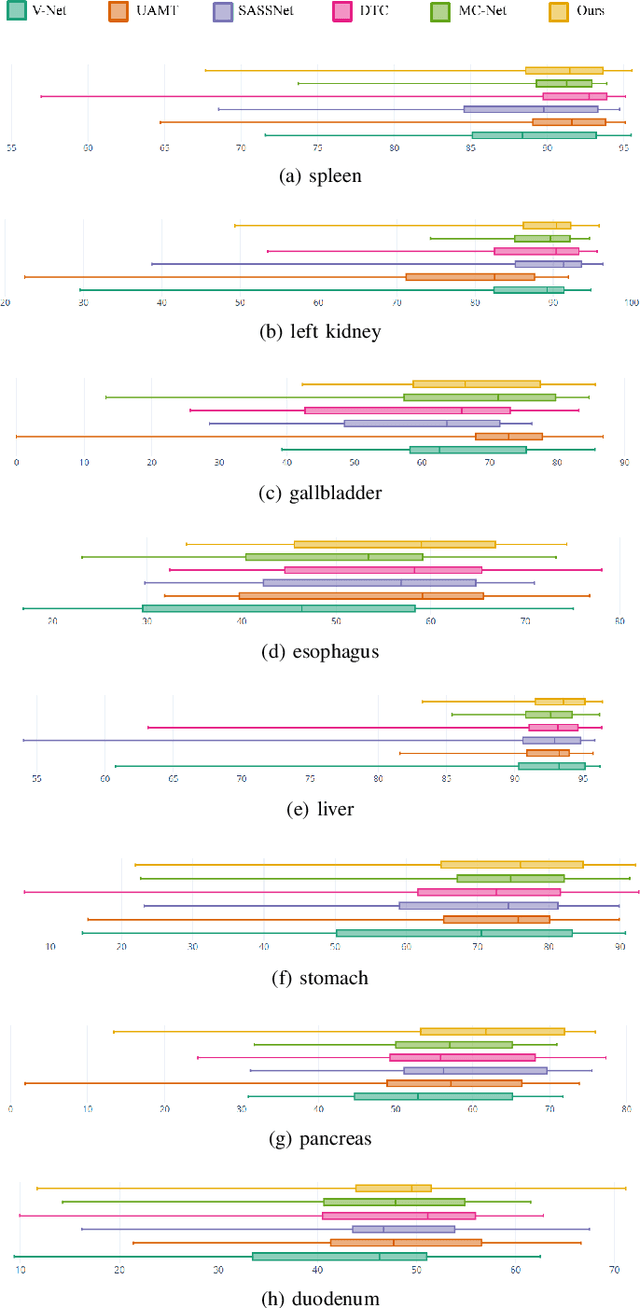
Abstract:Semi-supervised learning for medical image segmentation is an important area of research for alleviating the huge cost associated with the construction of reliable large-scale annotations in the medical domain. Recent semi-supervised approaches have demonstrated promising results by employing consistency regularization, pseudo-labeling techniques, and adversarial learning. These methods primarily attempt to learn the distribution of labeled and unlabeled data by enforcing consistency in the predictions or embedding context. However, previous approaches have focused only on local discrepancy minimization or context relations across single classes. In this paper, we introduce a novel adversarial learning-based semi-supervised segmentation method that effectively embeds both local and global features from multiple hidden layers and learns context relations between multiple classes. Our voxel-wise adversarial learning method utilizes a voxel-wise feature discriminator, which considers multilayer voxel-wise features (involving both local and global features) as an input by embedding class-specific voxel-wise feature distribution. Furthermore, we improve our previous representation learning method by overcoming information loss and learning stability problems, which enables rich representations of labeled data. Our method outperforms current best-performing state-of-the-art semi-supervised learning approaches on the image segmentation of the left atrium (single class) and multiorgan datasets (multiclass). Moreover, our visual interpretation of the feature space demonstrates that our proposed method enables a well-distributed and separated feature space from both labeled and unlabeled data, which improves the overall prediction results.
Voxel-level Siamese Representation Learning for Abdominal Multi-Organ Segmentation
May 17, 2021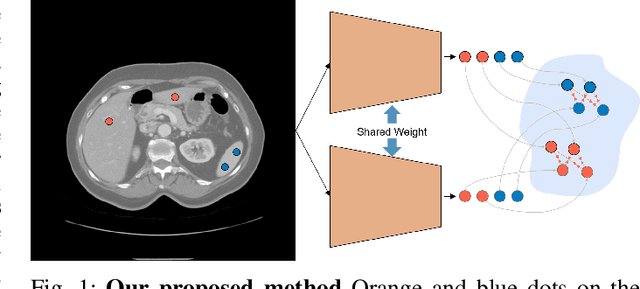
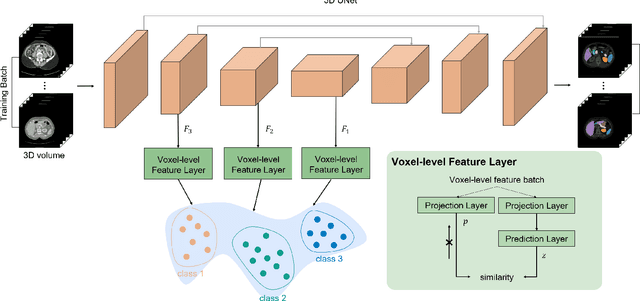
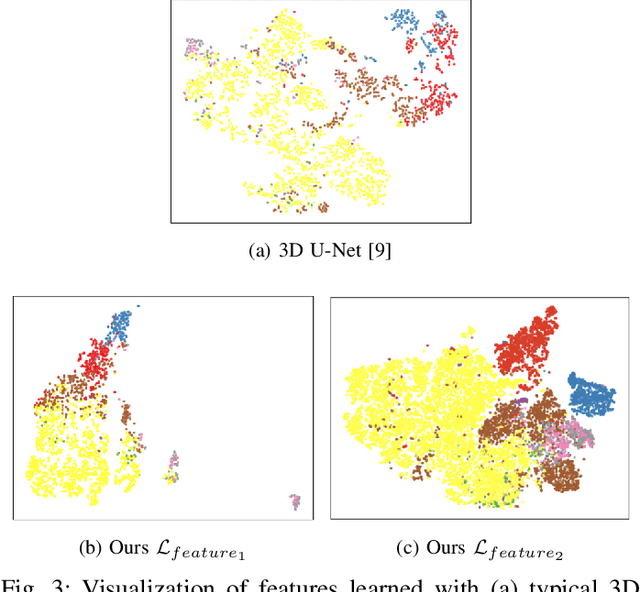
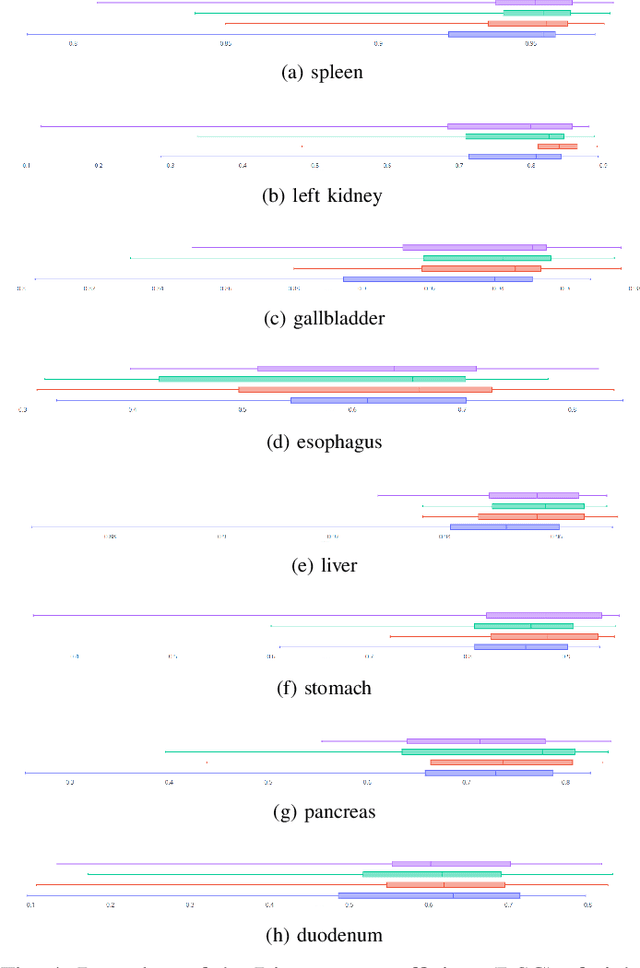
Abstract:Recent works in medical image segmentation have actively explored various deep learning architectures or objective functions to encode high-level features from volumetric data owing to limited image annotations. However, most existing approaches tend to ignore cross-volume global context and define context relations in the decision space. In this work, we propose a novel voxel-level Siamese representation learning method for abdominal multi-organ segmentation to improve representation space. The proposed method enforces voxel-wise feature relations in the representation space for leveraging limited datasets more comprehensively to achieve better performance. Inspired by recent progress in contrastive learning, we suppressed voxel-wise relations from the same class to be projected to the same point without using negative samples. Moreover, we introduce a multi-resolution context aggregation method that aggregates features from multiple hidden layers, which encodes both the global and local contexts for segmentation. Our experiments on the multi-organ dataset outperformed the existing approaches by 2% in Dice score coefficient. The qualitative visualizations of the representation spaces demonstrate that the improvements were gained primarily by a disentangled feature space.
Individual Tooth Detection and Identification from Dental Panoramic X-Ray Images via Point-wise Localization and Distance Regularization
Apr 12, 2020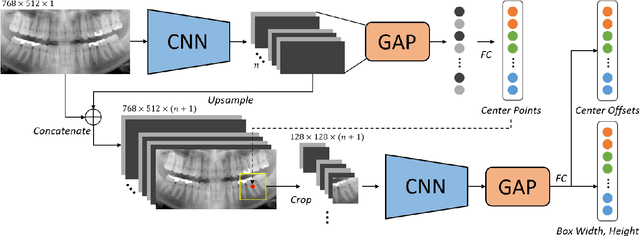
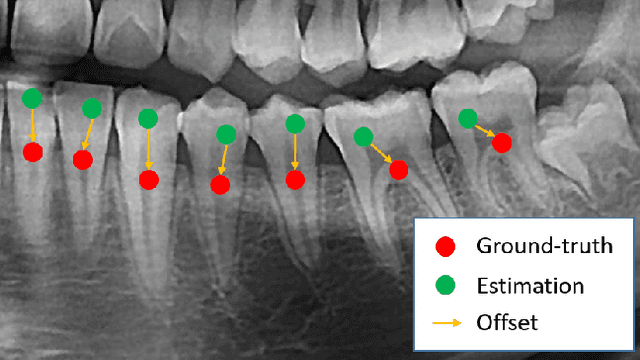
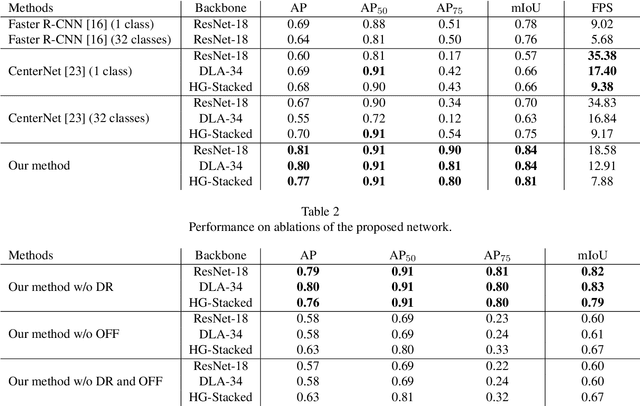
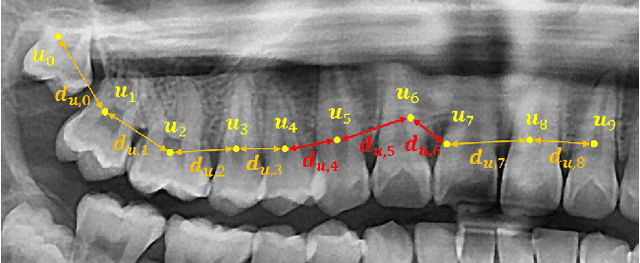
Abstract:Dental panoramic X-ray imaging is a popular diagnostic method owing to its very small dose of radiation. For an automated computer-aided diagnosis system in dental clinics, automatic detection and identification of individual teeth from panoramic X-ray images are critical prerequisites. In this study, we propose a point-wise tooth localization neural network by introducing a spatial distance regularization loss. The proposed network initially performs center point regression for all the anatomical teeth (i.e., 32 points), which automatically identifies each tooth. A novel distance regularization penalty is employed on the 32 points by considering $L_2$ regularization loss of Laplacian on spatial distances. Subsequently, teeth boxes are individually localized using a cascaded neural network on a patch basis. A multitask offset training is employed on the final output to improve the localization accuracy. Our method successfully localizes not only the existing teeth but also missing teeth; consequently, highly accurate detection and identification are achieved. The experimental results demonstrate that the proposed algorithm outperforms state-of-the-art approaches by increasing the average precision of teeth detection by 15.71% compared to the best performing method. The accuracy of identification achieved a precision of 0.997 and recall value of 0.972. Moreover, the proposed network does not require any additional identification algorithm owing to the preceding regression of the fixed 32 points regardless of the existence of the teeth.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge