Catalina Gomez
Intelligent Control of Robotic X-ray Devices using a Language-promptable Digital Twin
Dec 11, 2024


Abstract:Natural language offers a convenient, flexible interface for controlling robotic C-arm X-ray systems, making advanced functionality and controls accessible. However, enabling language interfaces requires specialized AI models that interpret X-ray images to create a semantic representation for reasoning. The fixed outputs of such AI models limit the functionality of language controls. Incorporating flexible, language-aligned AI models prompted through language enables more versatile interfaces for diverse tasks and procedures. Using a language-aligned foundation model for X-ray image segmentation, our system continually updates a patient digital twin based on sparse reconstructions of desired anatomical structures. This supports autonomous capabilities such as visualization, patient-specific viewfinding, and automatic collimation from novel viewpoints, enabling commands 'Focus in on the lower lumbar vertebrae.' In a cadaver study, users visualized, localized, and collimated structures across the torso using verbal commands, achieving 84% end-to-end success. Post hoc analysis of randomly oriented images showed our patient digital twin could localize 35 commonly requested structures to within 51.68 mm, enabling localization and isolation from arbitrary orientations. Our results demonstrate how intelligent robotic X-ray systems can incorporate physicians' expressed intent directly. While existing foundation models for intra-operative X-ray analysis exhibit failure modes, as they improve, they can facilitate highly flexible, intelligent robotic C-arms.
Designing AI Support for Human Involvement in AI-assisted Decision Making: A Taxonomy of Human-AI Interactions from a Systematic Review
Oct 31, 2023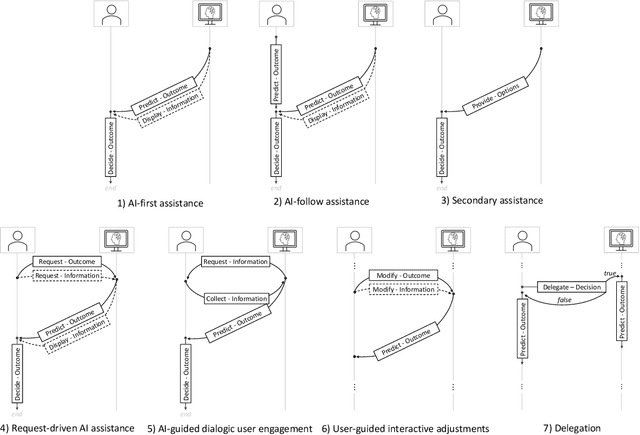

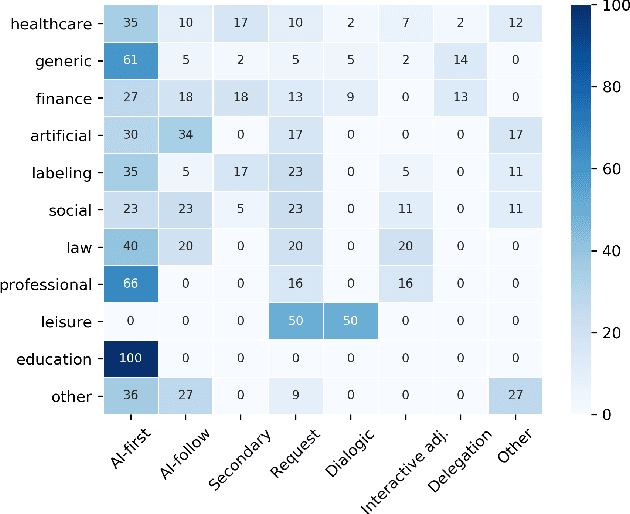

Abstract:Efforts in levering Artificial Intelligence (AI) in decision support systems have disproportionately focused on technological advancements, often overlooking the alignment between algorithmic outputs and human expectations. To address this, explainable AI promotes AI development from a more human-centered perspective. Determining what information AI should provide to aid humans is vital, however, how the information is presented, e. g., the sequence of recommendations and the solicitation of interpretations, is equally crucial. This motivates the need to more precisely study Human-AI interaction as a pivotal component of AI-based decision support. While several empirical studies have evaluated Human-AI interactions in multiple application domains in which interactions can take many forms, there is not yet a common vocabulary to describe human-AI interaction protocols. To address this gap, we describe the results of a systematic review of the AI-assisted decision making literature, analyzing 105 selected articles, which grounds the introduction of a taxonomy of interaction patterns that delineate various modes of human-AI interactivity. We find that current interactions are dominated by simplistic collaboration paradigms and report comparatively little support for truly interactive functionality. Our taxonomy serves as a valuable tool to understand how interactivity with AI is currently supported in decision-making contexts and foster deliberate choices of interaction designs.
INTRPRT: A Systematic Review of and Guidelines for Designing and Validating Transparent AI in Medical Image Analysis
Dec 21, 2021
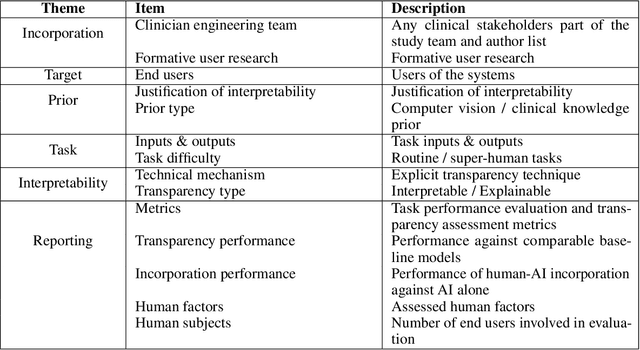

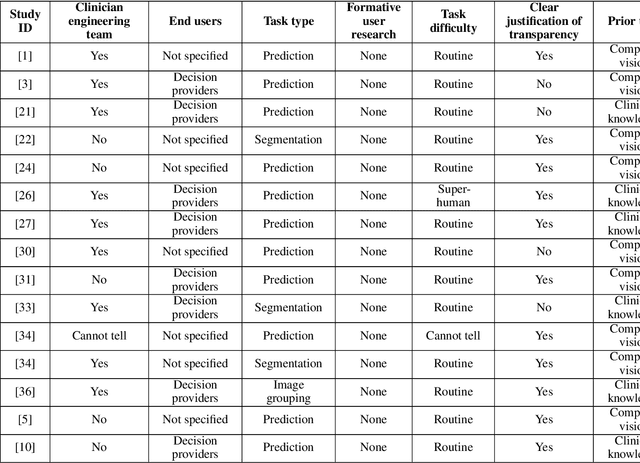
Abstract:Transparency in Machine Learning (ML), attempts to reveal the working mechanisms of complex models. Transparent ML promises to advance human factors engineering goals of human-centered AI in the target users. From a human-centered design perspective, transparency is not a property of the ML model but an affordance, i.e. a relationship between algorithm and user; as a result, iterative prototyping and evaluation with users is critical to attaining adequate solutions that afford transparency. However, following human-centered design principles in healthcare and medical image analysis is challenging due to the limited availability of and access to end users. To investigate the state of transparent ML in medical image analysis, we conducted a systematic review of the literature. Our review reveals multiple severe shortcomings in the design and validation of transparent ML for medical image analysis applications. We find that most studies to date approach transparency as a property of the model itself, similar to task performance, without considering end users during neither development nor evaluation. Additionally, the lack of user research, and the sporadic validation of transparency claims put contemporary research on transparent ML for medical image analysis at risk of being incomprehensible to users, and thus, clinically irrelevant. To alleviate these shortcomings in forthcoming research while acknowledging the challenges of human-centered design in healthcare, we introduce the INTRPRT guideline, a systematic design directive for transparent ML systems in medical image analysis. The INTRPRT guideline suggests formative user research as the first step of transparent model design to understand user needs and domain requirements. Following this process produces evidence to support design choices, and ultimately, increases the likelihood that the algorithms afford transparency.
An Interpretable Algorithm for Uveal Melanoma Subtyping from Whole Slide Cytology Images
Aug 13, 2021



Abstract:Algorithmic decision support is rapidly becoming a staple of personalized medicine, especially for high-stakes recommendations in which access to certain information can drastically alter the course of treatment, and thus, patient outcome; a prominent example is radiomics for cancer subtyping. Because in these scenarios the stakes are high, it is desirable for decision systems to not only provide recommendations but supply transparent reasoning in support thereof. For learning-based systems, this can be achieved through an interpretable design of the inference pipeline. Herein we describe an automated yet interpretable system for uveal melanoma subtyping with digital cytology images from fine needle aspiration biopsies. Our method embeds every automatically segmented cell of a candidate cytology image as a point in a 2D manifold defined by many representative slides, which enables reasoning about the cell-level composition of the tissue sample, paving the way for interpretable subtyping of the biopsy. Finally, a rule-based slide-level classification algorithm is trained on the partitions of the circularly distorted 2D manifold. This process results in a simple rule set that is evaluated automatically but highly transparent for human verification. On our in house cytology dataset of 88 uveal melanoma patients, the proposed method achieves an accuracy of 87.5% that compares favorably to all competing approaches, including deep "black box" models. The method comes with a user interface to facilitate interaction with cell-level content, which may offer additional insights for pathological assessment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge