César Domínguez
Semi-Supervised Machine Learning: a Homological Approach
Jan 27, 2023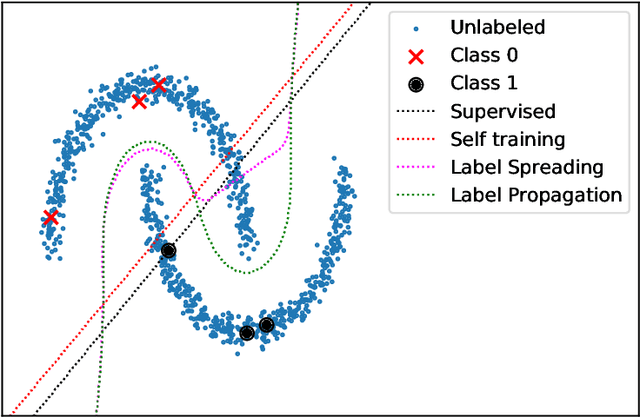
Abstract:In this paper we describe the mathematical foundations of a new approach to semi-supervised Machine Learning. Using techniques of Symbolic Computation and Computer Algebra, we apply the concept of persistent homology to obtain a new semi-supervised learning method.
Semi-Supervised Learning for Image Classification using Compact Networks in the BioMedical Context
May 19, 2022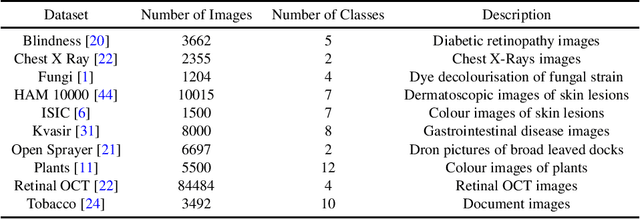

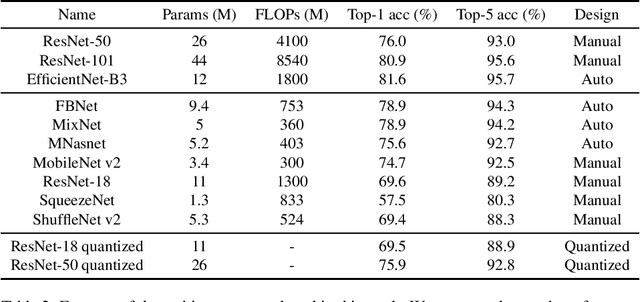
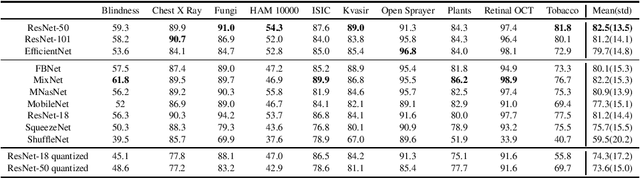
Abstract:The development of mobile and on the edge applications that embed deep convolutional neural models has the potential to revolutionise biomedicine. However, most deep learning models require computational resources that are not available in smartphones or edge devices; an issue that can be faced by means of compact models. The problem with such models is that they are, at least usually, less accurate than bigger models. In this work, we study how this limitation can be addressed with the application of semi-supervised learning techniques. We conduct several statistical analyses to compare performance of deep compact architectures when trained using semi-supervised learning methods for tackling image classification tasks in the biomedical context. In particular, we explore three families of compact networks, and two families of semi-supervised learning techniques for 10 biomedical tasks. By combining semi-supervised learning methods with compact networks, it is possible to obtain a similar performance to standard size networks. In general, the best results are obtained when combining data distillation with MixNet, and plain distillation with ResNet-18. Also, in general, NAS networks obtain better results than manually designed networks and quantized networks. The work presented in this paper shows the benefits of apply semi-supervised methods to compact networks; this allow us to create compact models that are not only as accurate as standard size models, but also faster and lighter. Finally, we have developed a library that simplifies the construction of compact models using semi-supervised learning methods.
A Topological Approach for Semi-Supervised Learning
May 19, 2022
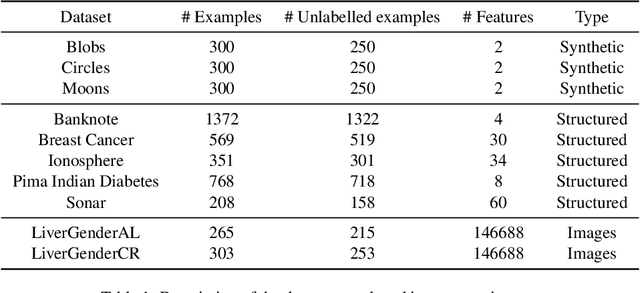
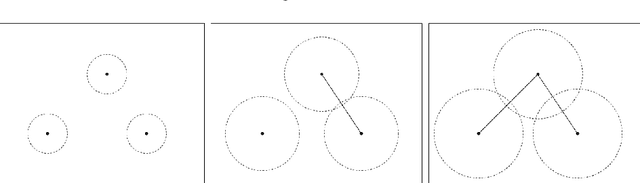
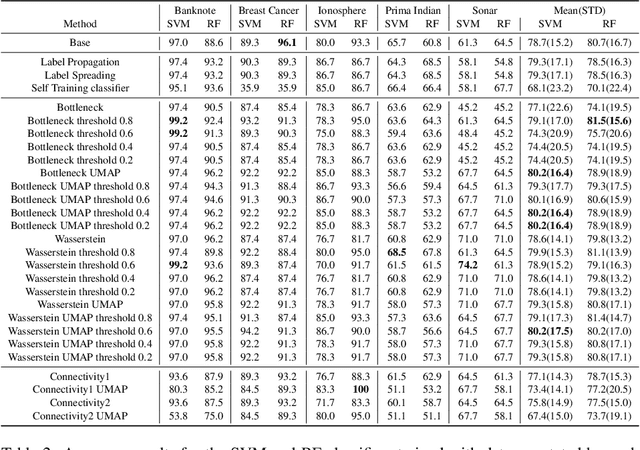
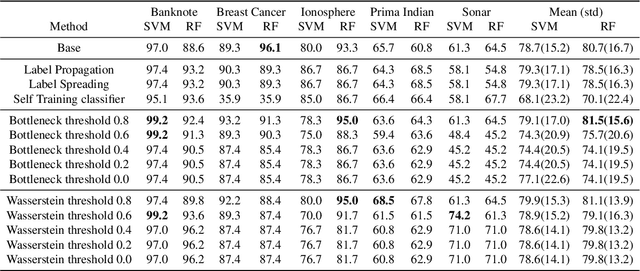
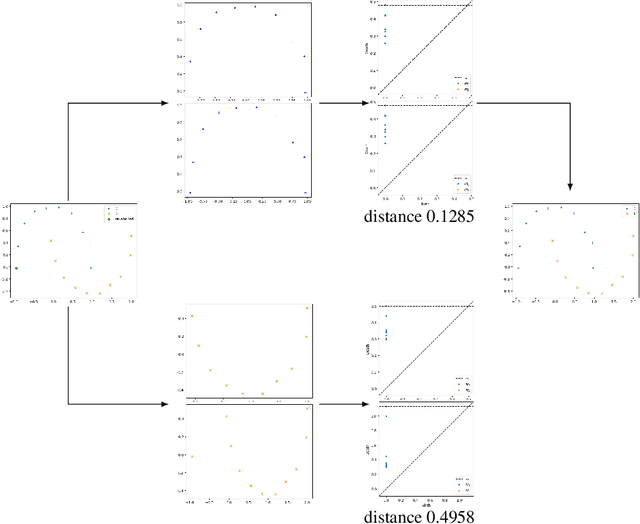
Abstract:Nowadays, Machine Learning and Deep Learning methods have become the state-of-the-art approach to solve data classification tasks. In order to use those methods, it is necessary to acquire and label a considerable amount of data; however, this is not straightforward in some fields, since data annotation is time consuming and might require expert knowledge. This challenge can be tackled by means of semi-supervised learning methods that take advantage of both labelled and unlabelled data. In this work, we present new semi-supervised learning methods based on techniques from Topological Data Analysis (TDA), a field that is gaining importance for analysing large amounts of data with high variety and dimensionality. In particular, we have created two semi-supervised learning methods following two different topological approaches. In the former, we have used a homological approach that consists in studying the persistence diagrams associated with the data using the Bottleneck and Wasserstein distances. In the latter, we have taken into account the connectivity of the data. In addition, we have carried out a thorough analysis of the developed methods using 3 synthetic datasets, 5 structured datasets, and 2 datasets of images. The results show that the semi-supervised methods developed in this work outperform both the results obtained with models trained with only manually labelled data, and those obtained with classical semi-supervised learning methods, reaching improvements of up to a 16%.
Neural Style Transfer and Unpaired Image-to-Image Translation to deal with the Domain Shift Problem on Spheroid Segmentation
Dec 16, 2021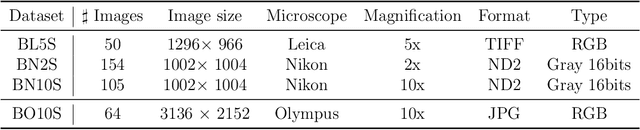
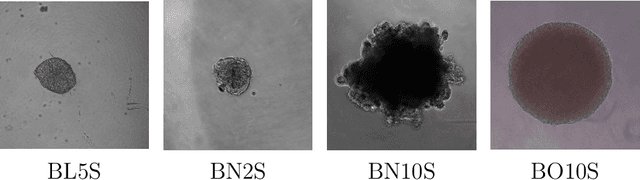
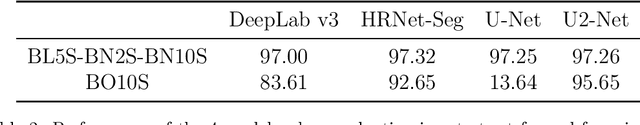
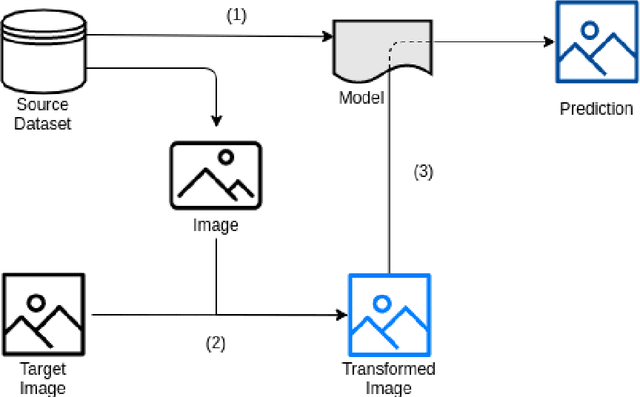
Abstract:Background and objectives. Domain shift is a generalisation problem of machine learning models that occurs when the data distribution of the training set is different to the data distribution encountered by the model when it is deployed. This is common in the context of biomedical image segmentation due to the variance of experimental conditions, equipment, and capturing settings. In this work, we address this challenge by studying both neural style transfer algorithms and unpaired image-to-image translation methods in the context of the segmentation of tumour spheroids. Methods. We have illustrated the domain shift problem in the context of spheroid segmentation with 4 deep learning segmentation models that achieved an IoU over 97% when tested with images following the training distribution, but whose performance decreased up to an 84\% when applied to images captured under different conditions. In order to deal with this problem, we have explored 3 style transfer algorithms (NST, deep image analogy, and STROTSS), and 6 unpaired image-to-image translations algorithms (CycleGAN, DualGAN, ForkGAN, GANILLA, CUT, and FastCUT). These algorithms have been integrated into a high-level API that facilitates their application to other contexts where the domain-shift problem occurs. Results. We have considerably improved the performance of the 4 segmentation models when applied to images captured under different conditions by using both style transfer and image-to-image translation algorithms. In particular, there are 2 style transfer algorithms (NST and deep image analogy) and 1 unpaired image-to-image translations algorithm (CycleGAN) that improve the IoU of the models in a range from 0.24 to 76.07. Therefore, reaching a similar performance to the one obtained with the models are applied to images following the training distribution.
Text Classification Models for Form Entity Linking
Dec 14, 2021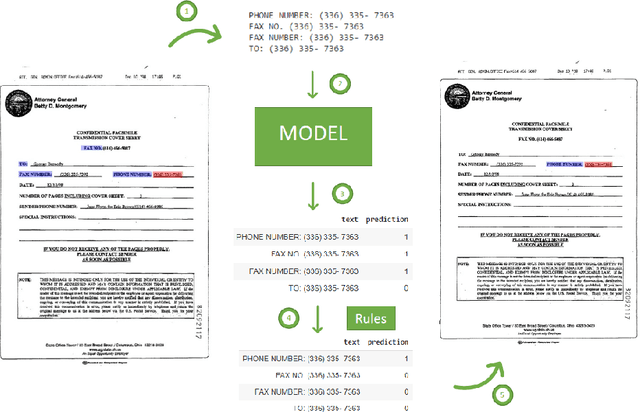
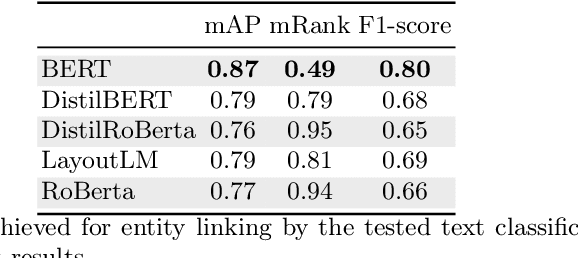
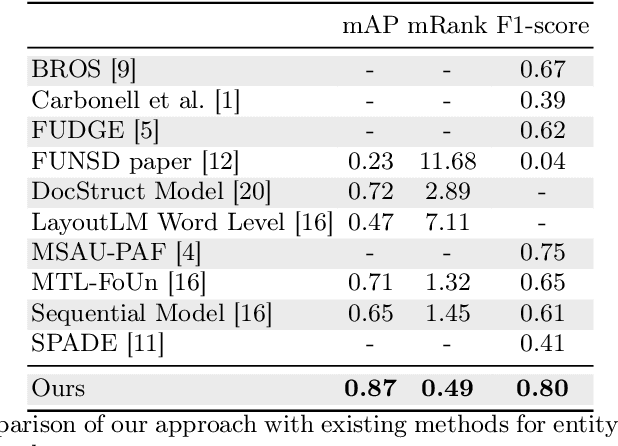
Abstract:Forms are a widespread type of template-based document used in a great variety of fields including, among others, administration, medicine, finance, or insurance. The automatic extraction of the information included in these documents is greatly demanded due to the increasing volume of forms that are generated in a daily basis. However, this is not a straightforward task when working with scanned forms because of the great diversity of templates with different location of form entities, and the quality of the scanned documents. In this context, there is a feature that is shared by all forms: they contain a collection of interlinked entities built as key-value (or label-value) pairs, together with other entities such as headers or images. In this work, we have tacked the problem of entity linking in forms by combining image processing techniques and a text classification model based on the BERT architecture. This approach achieves state-of-the-art results with a F1-score of 0.80 on the FUNSD dataset, a 5% improvement regarding the best previous method. The code of this project is available at https://github.com/mavillot/FUNSD-Entity-Linking.
The Benefits of Close-Domain Fine-Tuning for Table Detection in Document Images
Dec 12, 2019
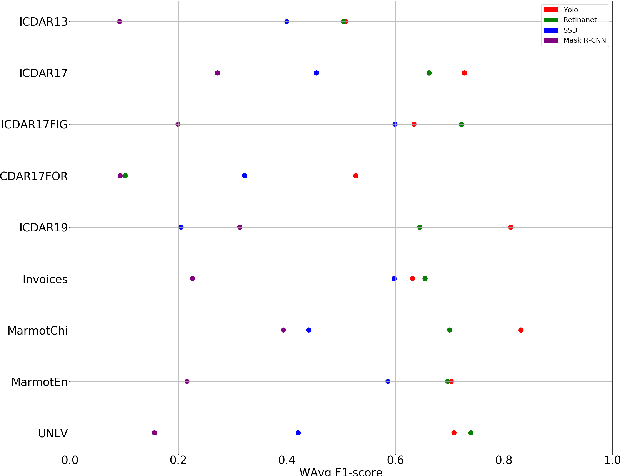
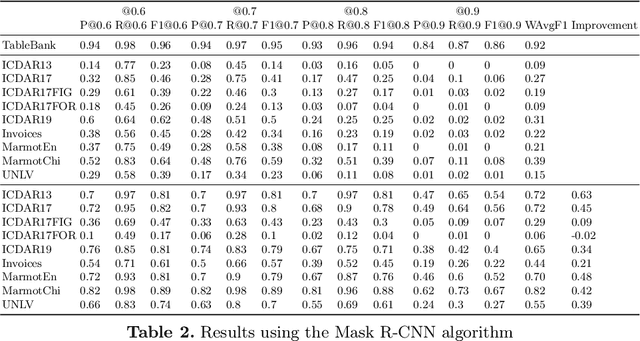
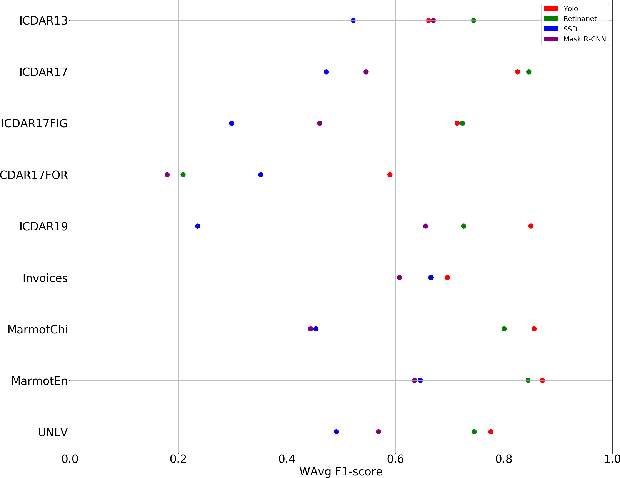
Abstract:A correct localisation of tables in a document is instrumental for determining their structure and extracting their contents; therefore, table detection is a key step in table understanding. Nowadays, the most successful methods for table detection in document images employ deep learning algorithms; and, particularly, a technique known as fine-tuning. In this context, such a technique exports the knowledge acquired to detect objects in natural images to detect tables in document images. However, there is only a vague relation between natural and document images, and fine-tuning works better when there is a close relation between the source and target task. In this paper, we show that it is more beneficial to employ fine-tuning from a closer domain. To this aim, we train different object detection algorithms (namely, Mask R-CNN, RetinaNet, SSD and YOLO) using the TableBank dataset (a dataset of images of academic documents designed for table detection and recognition), and fine-tune them for several heterogeneous table detection datasets. Using this approach, we considerably improve the accuracy of the detection models fine-tuned from natural images (in mean a 17%, and, in the best case, up to a 60%).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge