Brighter Agyemang
Distributed Interaction Graph Construction for Dynamic DCOPs in Cooperative Multi-agent Systems
Dec 07, 2022


Abstract:DCOP algorithms usually rely on interaction graphs to operate. In open and dynamic environments, such methods need to address how this interaction graph is generated and maintained among agents. Existing methods require reconstructing the entire graph upon detecting changes in the environment or assuming that new agents know potential neighbors to facilitate connection. We propose a novel distributed interaction graph construction algorithm to address this problem. The proposed method does not assume a predefined constraint graph and stabilizes after disruptive changes in the environment. We evaluate our approach by pairing it with existing DCOP algorithms to solve several generated dynamic problems. The experiment results show that the proposed algorithm effectively constructs and maintains a stable multi-agent interaction graph for open and dynamic environments.
Deep Inverse Reinforcement Learning for Structural Evolution of Small Molecules
Jul 24, 2020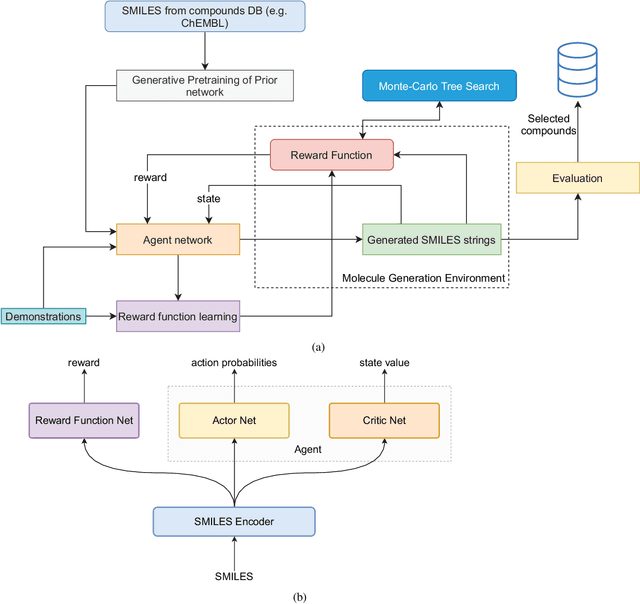
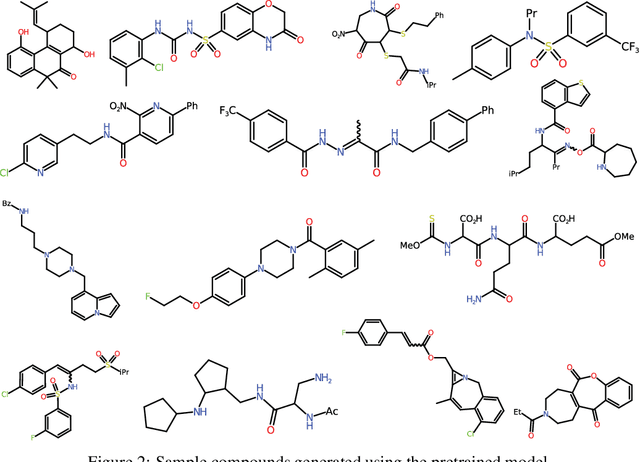
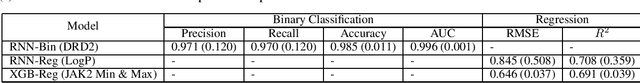
Abstract:The size and quality of chemical libraries to the drug discovery pipeline are crucial for developing new drugs or repurposing existing drugs. Existing techniques such as combinatorial organic synthesis and High-Throughput Screening usually make the process extraordinarily tough and complicated since the search space of synthetically feasible drugs is exorbitantly huge. While reinforcement learning has been mostly exploited in the literature for generating novel compounds, the requirement of designing a reward function that succinctly represents the learning objective could prove daunting in certain complex domains. Generative Adversarial Network-based methods also mostly discard the discriminator after training and could be hard to train. In this study, we propose a framework for training a compound generator and learning a transferable reward function based on the entropy maximization inverse reinforcement learning paradigm. We show from our experiments that the inverse reinforcement learning route offers a rational alternative for generating chemical compounds in domains where reward function engineering may be less appealing or impossible while data exhibiting the desired objective is readily available.
Multi-View Self-Attention for Interpretable Drug-Target Interaction Prediction
May 01, 2020



Abstract:The drug discovery stage is a vital part of the drug development process and forms part of the initial stages of the development pipeline. In recent times, machine learning-based methods are actively being used to model drug-target interactions for rational drug discovery due to the successful application of these methods in other domains. In machine learning approaches, the numerical representation of molecules is vital to the performance of the model. While significant progress has been made in molecular representation engineering, this has resulted in several descriptors for both targets and compounds. Also, the interpretability of model predictions is a vital feature that could have several pharmacological applications. In this study, we propose a self-attention-based, multi-view representation learning approach for modeling drug-target interactions. We evaluated our approach using three large-scale kinase datasets and compared six variants of our method to 16 baselines. Our experimental results demonstrate the ability of our method to achieve high accuracy and offer biologically plausible interpretations using neural attention.
Drug-Target Indication Prediction by Integrating End-to-End Learning and Fingerprints
Dec 06, 2019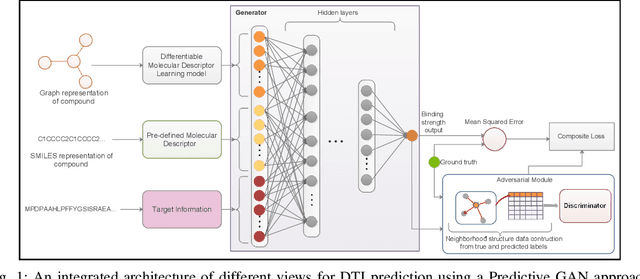
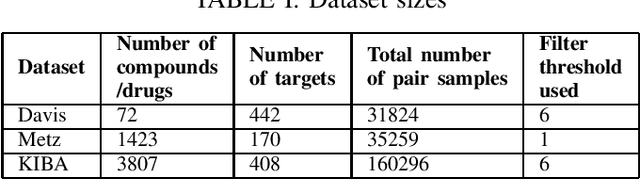
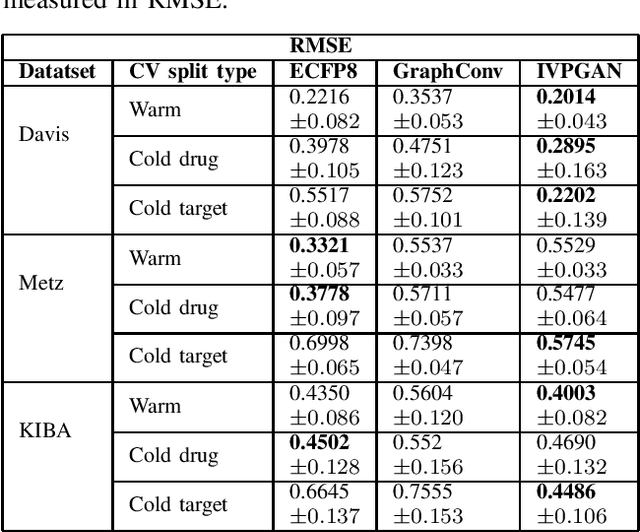
Abstract:Computer-Aided Drug Discovery research has proven to be a promising direction in drug discovery. In recent years, Deep Learning approaches have been applied to problems in the domain such as Drug-Target Interaction Prediction and have shown improvements over traditional screening methods. An existing challenge is how to represent compound-target pairs in deep learning models. While several representation methods exist, such descriptor schemes tend to complement one another in many instances, as reported in the literature. In this study, we propose a multi-view architecture trained adversarially to leverage this complementary behavior by integrating both differentiable and predefined molecular descriptors. We conduct experiments on clinically relevant benchmark datasets to demonstrate the potential of our approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge