Brent Weinberg
MD, PhD
The 2024 Brain Tumor Segmentation (BraTS) Challenge: Glioma Segmentation on Post-treatment MRI
May 28, 2024
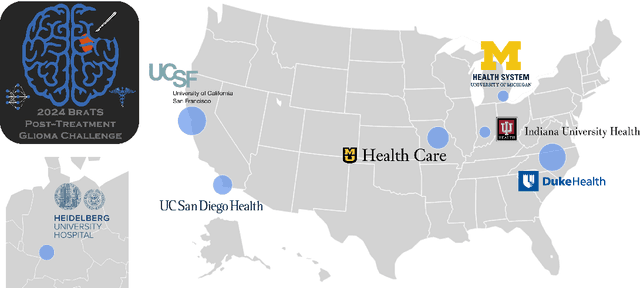
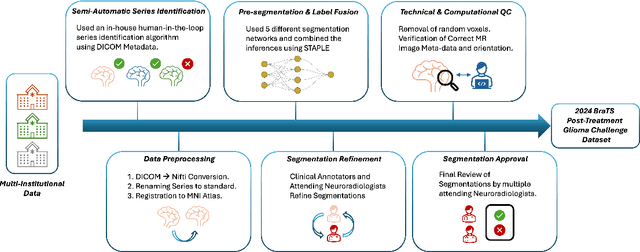
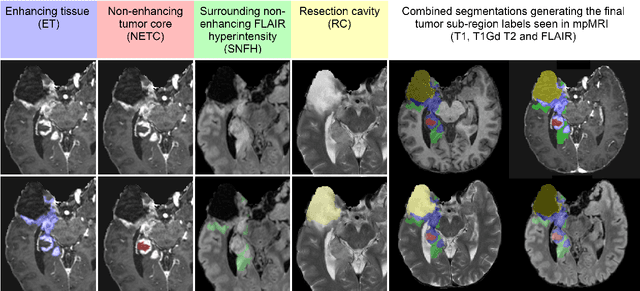
Abstract:Gliomas are the most common malignant primary brain tumors in adults and one of the deadliest types of cancer. There are many challenges in treatment and monitoring due to the genetic diversity and high intrinsic heterogeneity in appearance, shape, histology, and treatment response. Treatments include surgery, radiation, and systemic therapies, with magnetic resonance imaging (MRI) playing a key role in treatment planning and post-treatment longitudinal assessment. The 2024 Brain Tumor Segmentation (BraTS) challenge on post-treatment glioma MRI will provide a community standard and benchmark for state-of-the-art automated segmentation models based on the largest expert-annotated post-treatment glioma MRI dataset. Challenge competitors will develop automated segmentation models to predict four distinct tumor sub-regions consisting of enhancing tissue (ET), surrounding non-enhancing T2/fluid-attenuated inversion recovery (FLAIR) hyperintensity (SNFH), non-enhancing tumor core (NETC), and resection cavity (RC). Models will be evaluated on separate validation and test datasets using standardized performance metrics utilized across the BraTS 2024 cluster of challenges, including lesion-wise Dice Similarity Coefficient and Hausdorff Distance. Models developed during this challenge will advance the field of automated MRI segmentation and contribute to their integration into clinical practice, ultimately enhancing patient care.
SAGE: Sequential Attribute Generator for Analyzing Glioblastomas using Limited Dataset
May 14, 2020



Abstract:While deep learning approaches have shown remarkable performance in many imaging tasks, most of these methods rely on availability of large quantities of data. Medical image data, however, is scarce and fragmented. Generative Adversarial Networks (GANs) have recently been very effective in handling such datasets by generating more data. If the datasets are very small, however, GANs cannot learn the data distribution properly, resulting in less diverse or low-quality results. One such limited dataset is that for the concurrent gain of 19 and 20 chromosomes (19/20 co-gain), a mutation with positive prognostic value in Glioblastomas (GBM). In this paper, we detect imaging biomarkers for the mutation to streamline the extensive and invasive prognosis pipeline. Since this mutation is relatively rare, i.e. small dataset, we propose a novel generative framework - the Sequential Attribute GEnerator (SAGE), that generates detailed tumor imaging features while learning from a limited dataset. Experiments show that not only does SAGE generate high quality tumors when compared to standard Deep Convolutional GAN (DC-GAN) and Wasserstein GAN with Gradient Penalty (WGAN-GP), it also captures the imaging biomarkers accurately.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge