Birgit Schoeberl
Tensor clustering with algebraic constraints gives interpretable groups of crosstalk mechanisms in breast cancer
Apr 28, 2017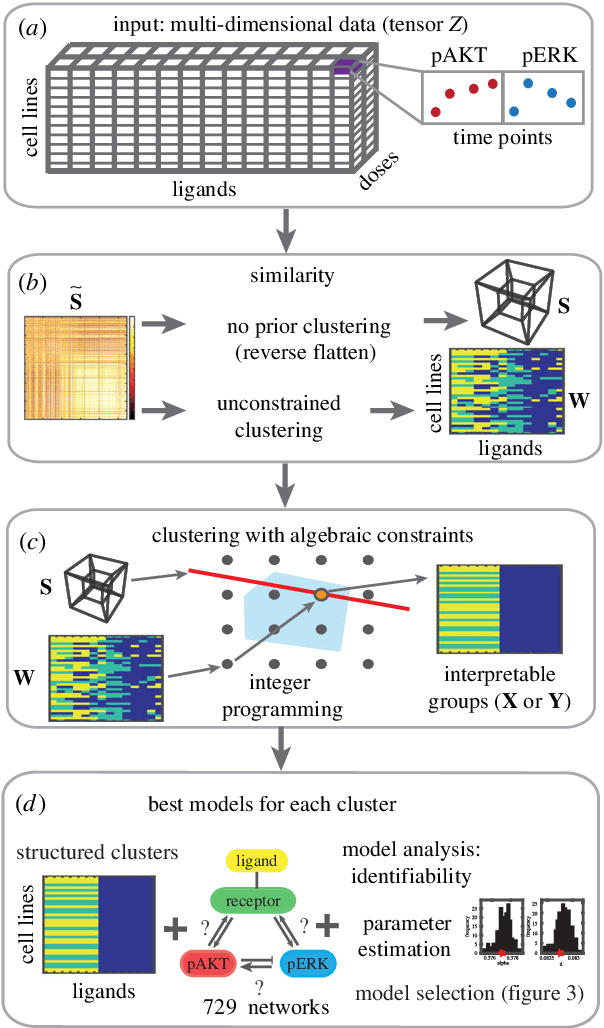
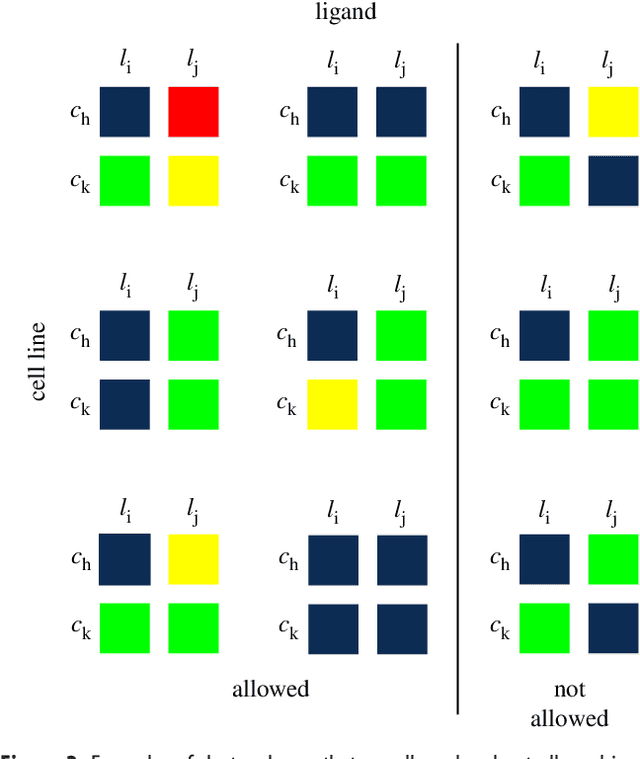
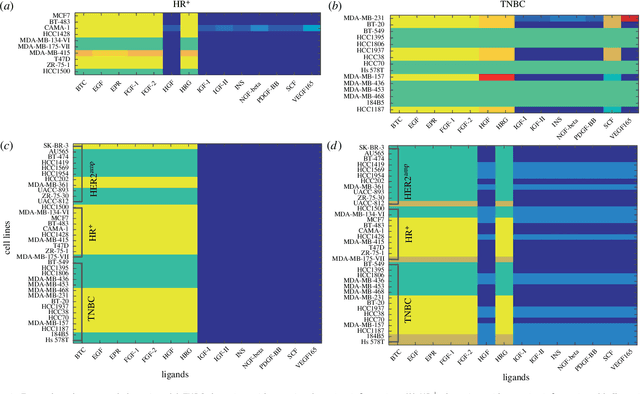
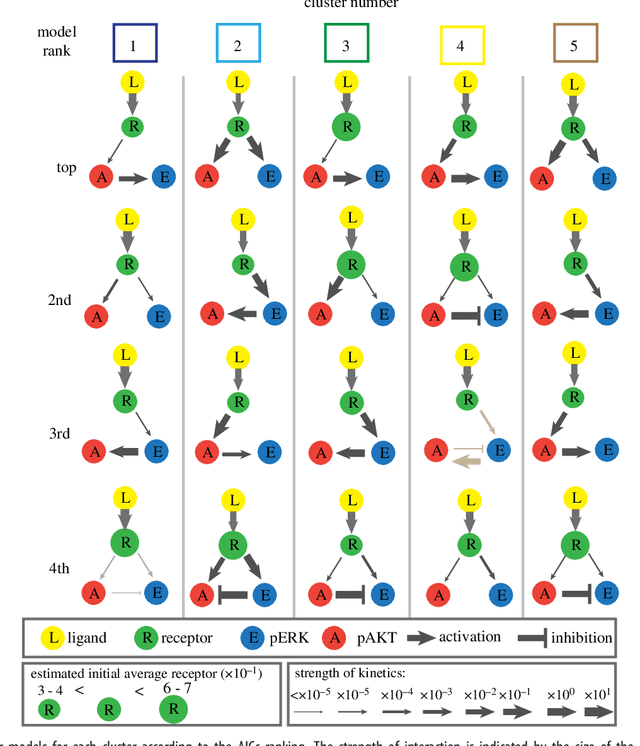
Abstract:We introduce a tensor-based clustering method to extract sparse, low-dimensional structure from high-dimensional, multi-indexed datasets. Specifically, this framework is designed to enable detection of clusters of data in the presence of structural requirements which we encode as algebraic constraints in a linear program. We illustrate our method on a collection of experiments measuring the response of genetically diverse breast cancer cell lines to an array of ligands. Each experiment consists of a cell line-ligand combination, and contains time-course measurements of the early-signalling kinases MAPK and AKT at two different ligand dose levels. By imposing appropriate structural constraints and respecting the multi-indexed structure of the data, our clustering analysis can be optimized for biological interpretation and therapeutic understanding. We then perform a systematic, large-scale exploration of mechanistic models of MAPK-AKT crosstalk for each cluster. This analysis allows us to quantify the heterogeneity of breast cancer cell subtypes, and leads to hypotheses about the mechanisms by which cell lines respond to ligands. Our clustering method is general and can be tailored to a variety of applications in science and industry.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge