Bernhard Y. Renard
BaGGLS: A Bayesian Shrinkage Framework for Interpretable Modeling of Interactions in High-Dimensional Biological Data
Nov 19, 2025



Abstract:Biological data sets are often high-dimensional, noisy, and governed by complex interactions among sparse signals. This poses major challenges for interpretability and reliable feature selection. Tasks such as identifying motif interactions in genomics exemplify these difficulties, as only a small subset of biologically relevant features (e.g., motifs) are typically active, and their effects are often non-linear and context-dependent. While statistical approaches often result in more interpretable models, deep learning models have proven effective in modeling complex interactions and prediction accuracy, yet their black-box nature limits interpretability. We introduce BaGGLS, a flexible and interpretable probabilistic binary regression model designed for high-dimensional biological inference involving feature interactions. BaGGLS incorporates a Bayesian group global-local shrinkage prior, aligned with the group structure introduced by interaction terms. This prior encourages sparsity while retaining interpretability, helping to isolate meaningful signals and suppress noise. To enable scalable inference, we employ a partially factorized variational approximation that captures posterior skewness and supports efficient learning even in large feature spaces. In extensive simulations, we can show that BaGGLS outperforms the other methods with regard to interaction detection and is many times faster than MCMC sampling under the horseshoe prior. We also demonstrate the usefulness of BaGGLS in the context of interaction discovery from motif scanner outputs and noisy attribution scores from deep learning models. This shows that BaGGLS is a promising approach for uncovering biologically relevant interaction patterns, with potential applicability across a range of high-dimensional tasks in computational biology.
Sparse Explanations of Neural Networks Using Pruned Layer-Wise Relevance Propagation
Apr 22, 2024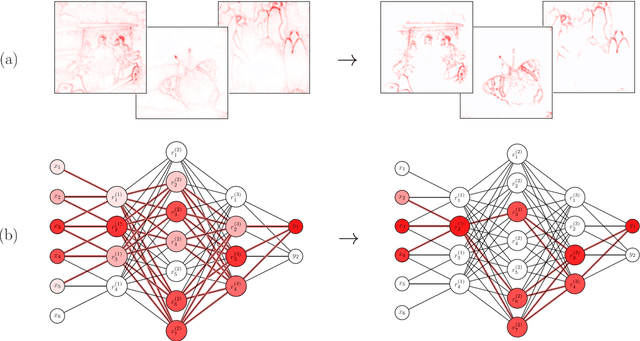
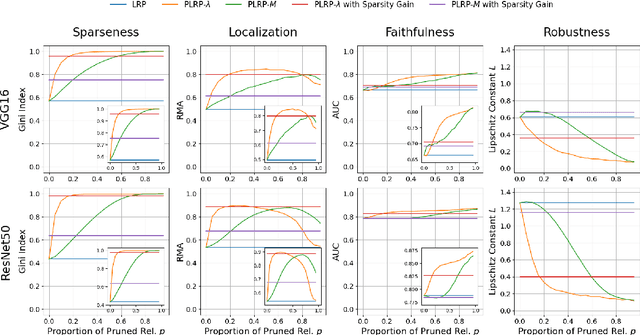
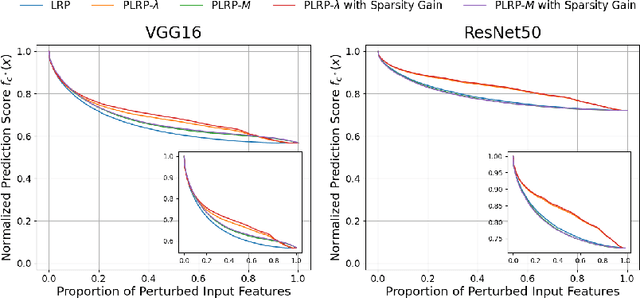
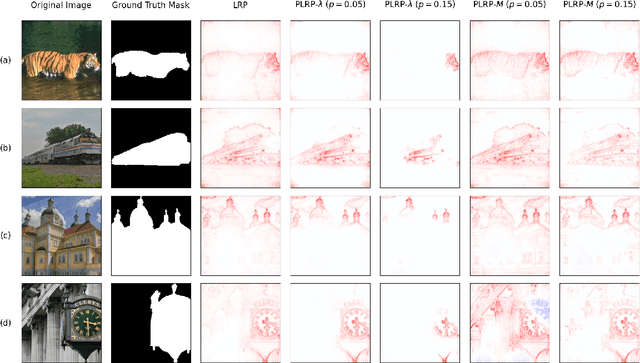
Abstract:Explainability is a key component in many applications involving deep neural networks (DNNs). However, current explanation methods for DNNs commonly leave it to the human observer to distinguish relevant explanations from spurious noise. This is not feasible anymore when going from easily human-accessible data such as images to more complex data such as genome sequences. To facilitate the accessibility of DNN outputs from such complex data and to increase explainability, we present a modification of the widely used explanation method layer-wise relevance propagation. Our approach enforces sparsity directly by pruning the relevance propagation for the different layers. Thereby, we achieve sparser relevance attributions for the input features as well as for the intermediate layers. As the relevance propagation is input-specific, we aim to prune the relevance propagation rather than the underlying model architecture. This allows to prune different neurons for different inputs and hence, might be more appropriate to the local nature of explanation methods. To demonstrate the efficacy of our method, we evaluate it on two types of data, images and genomic sequences. We show that our modification indeed leads to noise reduction and concentrates relevance on the most important features compared to the baseline.
Identifying Drivers of Predictive Uncertainty using Variance Feature Attribution
Dec 12, 2023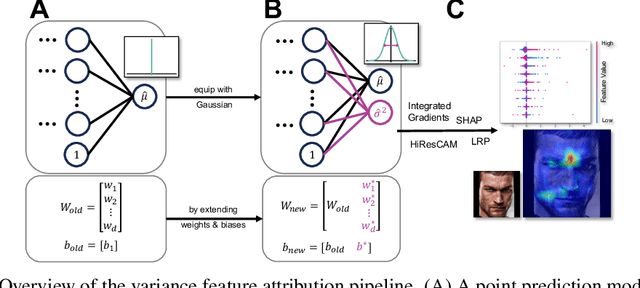
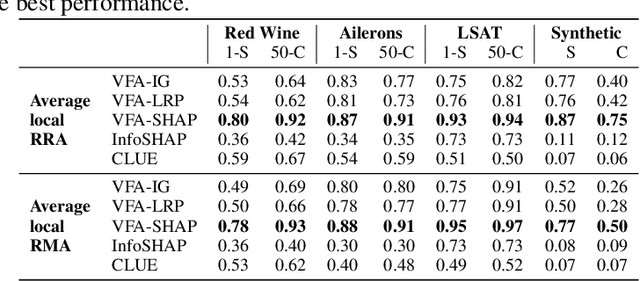
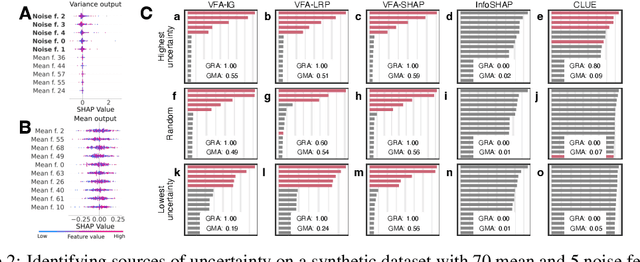

Abstract:Explainability and uncertainty quantification are two pillars of trustable artificial intelligence. However, the reasoning behind uncertainty estimates is generally left unexplained. Identifying the drivers of uncertainty complements explanations of point predictions in recognizing potential model limitations. It facilitates the detection of oversimplification in the uncertainty estimation process. Explanations of uncertainty enhance communication and trust in decisions. They allow for verifying whether the main drivers of model uncertainty are relevant and may impact model usage. So far, the subject of explaining uncertainties has been rarely studied. The few exceptions in existing literature are tailored to Bayesian neural networks or rely heavily on technically intricate approaches, hindering their broad adoption. We propose variance feature attribution, a simple and scalable solution to explain predictive aleatoric uncertainties. First, we estimate uncertainty as predictive variance by equipping a neural network with a Gaussian output distribution by adding a variance output neuron. Thereby, we can rely on pre-trained point prediction models and fine-tune them for meaningful variance estimation. Second, we apply out-of-the-box explainers on the variance output of these models to explain the uncertainty estimation. We evaluate our approach in a synthetic setting where the data-generating process is known. We show that our method can explain uncertainty influences more reliably and faster than the established baseline CLUE. We fine-tune a state-of-the-art age regression model to estimate uncertainty and obtain attributions. Our explanations highlight potential sources of uncertainty, such as laugh lines. Variance feature attribution provides accurate explanations for uncertainty estimates with little modifications to the model architecture and low computational overhead.
SimbaML: Connecting Mechanistic Models and Machine Learning with Augmented Data
Apr 08, 2023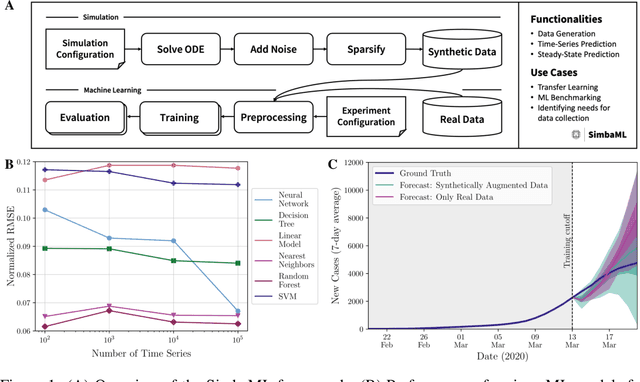
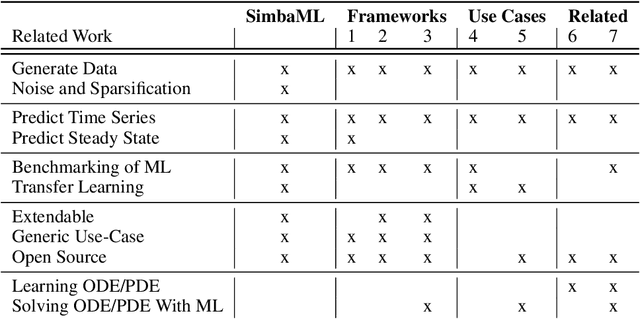
Abstract:Training sophisticated machine learning (ML) models requires large datasets that are difficult or expensive to collect for many applications. If prior knowledge about system dynamics is available, mechanistic representations can be used to supplement real-world data. We present SimbaML (Simulation-Based ML), an open-source tool that unifies realistic synthetic dataset generation from ordinary differential equation-based models and the direct analysis and inclusion in ML pipelines. SimbaML conveniently enables investigating transfer learning from synthetic to real-world data, data augmentation, identifying needs for data collection, and benchmarking physics-informed ML approaches. SimbaML is available from https://pypi.org/project/simba-ml/.
HiClass: a Python library for local hierarchical classification compatible with scikit-learn
Dec 20, 2021

Abstract:HiClass is an open-source Python package for local hierarchical classification fully compatible with scikit-learn. It provides implementations of the most popular machine learning models for local hierarchical classification, including Local Classifier Per Node, Local Classifier Per Parent Node and Local Classifier Per Level. In addition, the library includes tools to evaluate model performance on hierarchical data. The documentation contains installation instructions, interactive notebooks, and a complete description of the API. HiClass is distributed under the simplified BSD license, encouraging its use in both academic and commercial settings. Source code and documentation are available at https://gitlab.com/dacs-hpi/hiclass.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge