Ayman El-Baz
Autonomous Extraction of Gleason Patterns for Grading Prostate Cancer using Multi-Gigapixel Whole Slide Images
Nov 01, 2020
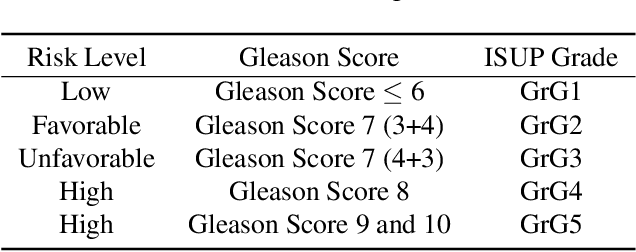
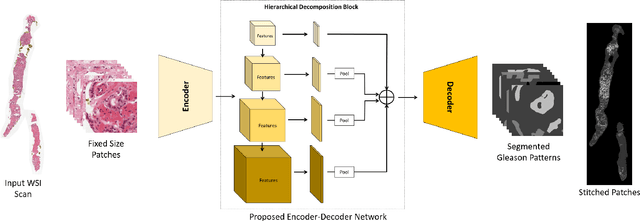
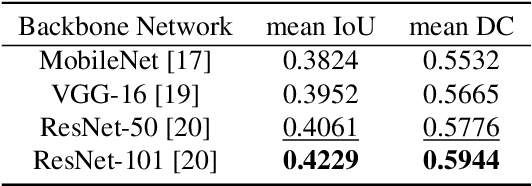
Abstract:Prostate cancer (PCa) is the second deadliest form of cancer in males. The severity of PCa can be clinically graded through the Gleason scores obtained by examining the structural representation of Gleason cellular patterns. This paper presents an asymmetric encoder-decoder model that integrates a novel hierarchical decomposition block to exploit the feature representations pooled across various scales and then fuses them together to generate the Gleason cellular patterns using the whole slide images. Furthermore, the proposed network is penalized through a novel three-tiered hybrid loss function which ensures that the proposed model accurately recognizes the cluttered regions of the cancerous tissues despite having similar contextual and textural characteristics. We have rigorously tested the proposed network on 10,516 whole slide scans (containing around 71.7M patches), where the proposed model achieved 3.59\% improvement over state-of-the-art scene parsing, encoder-decoder, and fully convolutional networks in terms of intersection-over-union.
Alzheimer's Disease Diagnostics by a Deeply Supervised Adaptable 3D Convolutional Network
Jul 02, 2016
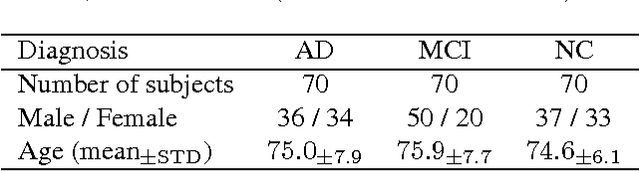
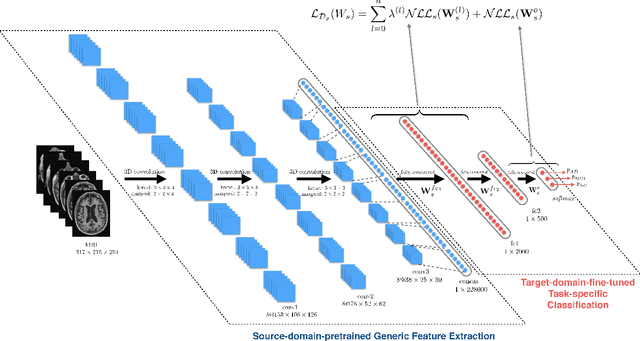
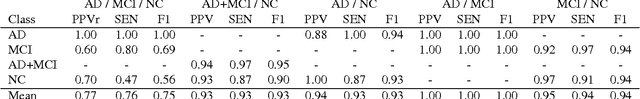
Abstract:Early diagnosis, playing an important role in preventing progress and treating the Alzheimer's disease (AD), is based on classification of features extracted from brain images. The features have to accurately capture main AD-related variations of anatomical brain structures, such as, e.g., ventricles size, hippocampus shape, cortical thickness, and brain volume. This paper proposes to predict the AD with a deep 3D convolutional neural network (3D-CNN), which can learn generic features capturing AD biomarkers and adapt to different domain datasets. The 3D-CNN is built upon a 3D convolutional autoencoder, which is pre-trained to capture anatomical shape variations in structural brain MRI scans. Fully connected upper layers of the 3D-CNN are then fine-tuned for each task-specific AD classification. Experiments on the \emph{ADNI} MRI dataset with no skull-stripping preprocessing have shown our 3D-CNN outperforms several conventional classifiers by accuracy and robustness. Abilities of the 3D-CNN to generalize the features learnt and adapt to other domains have been validated on the \emph{CADDementia} dataset.
Alzheimer's Disease Diagnostics by Adaptation of 3D Convolutional Network
Jul 02, 2016



Abstract:Early diagnosis, playing an important role in preventing progress and treating the Alzheimer\{'}s disease (AD), is based on classification of features extracted from brain images. The features have to accurately capture main AD-related variations of anatomical brain structures, such as, e.g., ventricles size, hippocampus shape, cortical thickness, and brain volume. This paper proposed to predict the AD with a deep 3D convolutional neural network (3D-CNN), which can learn generic features capturing AD biomarkers and adapt to different domain datasets. The 3D-CNN is built upon a 3D convolutional autoencoder, which is pre-trained to capture anatomical shape variations in structural brain MRI scans. Fully connected upper layers of the 3D-CNN are then fine-tuned for each task-specific AD classification. Experiments on the CADDementia MRI dataset with no skull-stripping preprocessing have shown our 3D-CNN outperforms several conventional classifiers by accuracy. Abilities of the 3D-CNN to generalize the features learnt and adapt to other domains have been validated on the ADNI dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge