Asheesh Singh
Agri-GNN: A Novel Genotypic-Topological Graph Neural Network Framework Built on GraphSAGE for Optimized Yield Prediction
Oct 19, 2023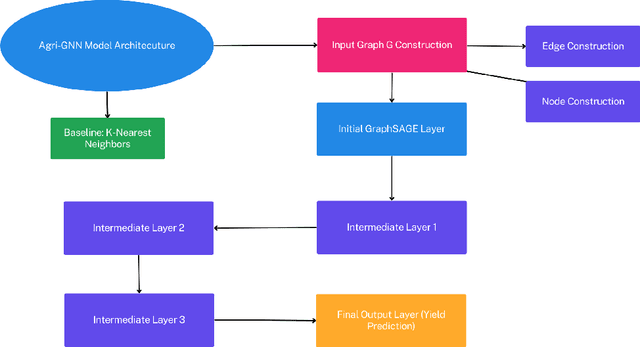

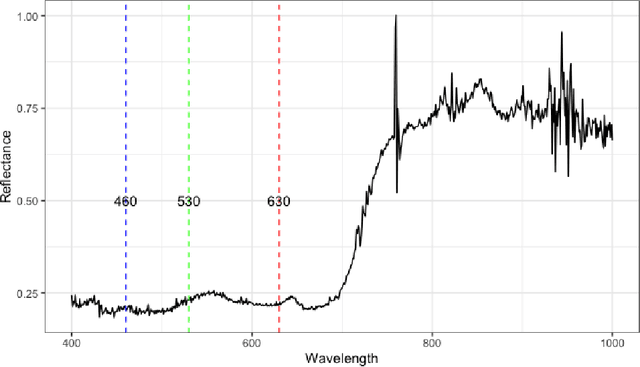
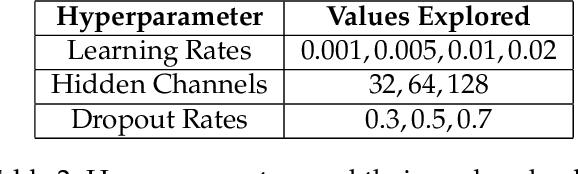
Abstract:Agriculture, as the cornerstone of human civilization, constantly seeks to integrate technology for enhanced productivity and sustainability. This paper introduces $\textit{Agri-GNN}$, a novel Genotypic-Topological Graph Neural Network Framework tailored to capture the intricate spatial and genotypic interactions of crops, paving the way for optimized predictions of harvest yields. $\textit{Agri-GNN}$ constructs a Graph $\mathcal{G}$ that considers farming plots as nodes, and then methodically constructs edges between nodes based on spatial and genotypic similarity, allowing for the aggregation of node information through a genotypic-topological filter. Graph Neural Networks (GNN), by design, consider the relationships between data points, enabling them to efficiently model the interconnected agricultural ecosystem. By harnessing the power of GNNs, $\textit{Agri-GNN}$ encapsulates both local and global information from plants, considering their inherent connections based on spatial proximity and shared genotypes, allowing stronger predictions to be made than traditional Machine Learning architectures. $\textit{Agri-GNN}$ is built from the GraphSAGE architecture, because of its optimal calibration with large graphs, like those of farming plots and breeding experiments. $\textit{Agri-GNN}$ experiments, conducted on a comprehensive dataset of vegetation indices, time, genotype information, and location data, demonstrate that $\textit{Agri-GNN}$ achieves an $R^2 = .876$ in yield predictions for farming fields in Iowa. The results show significant improvement over the baselines and other work in the field. $\textit{Agri-GNN}$ represents a blueprint for using advanced graph-based neural architectures to predict crop yield, providing significant improvements over baselines in the field.
An end-to-end convolutional selective autoencoder approach to Soybean Cyst Nematode eggs detection
Mar 25, 2016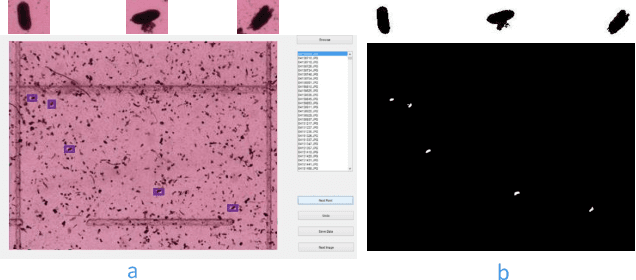
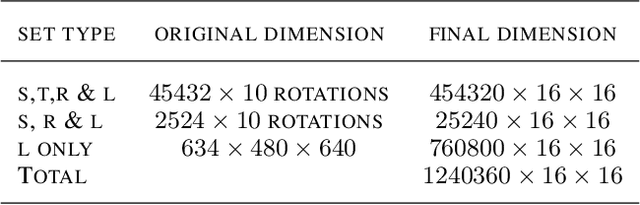
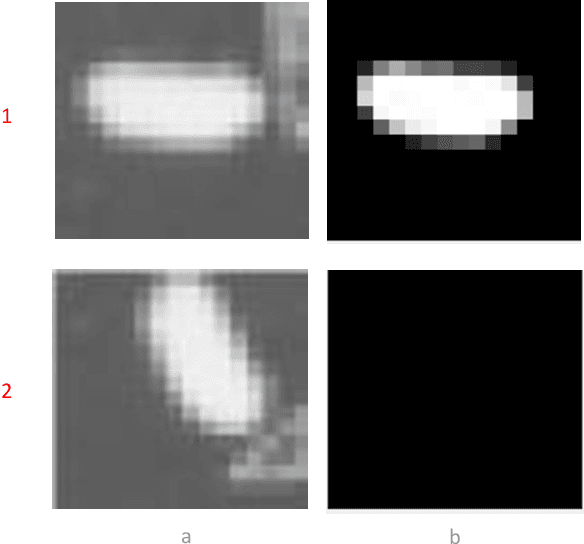
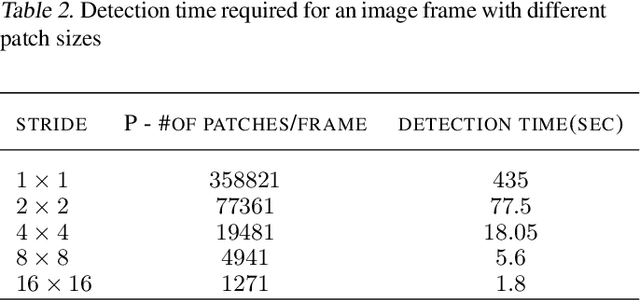
Abstract:This paper proposes a novel selective autoencoder approach within the framework of deep convolutional networks. The crux of the idea is to train a deep convolutional autoencoder to suppress undesired parts of an image frame while allowing the desired parts resulting in efficient object detection. The efficacy of the framework is demonstrated on a critical plant science problem. In the United States, approximately $1 billion is lost per annum due to a nematode infection on soybean plants. Currently, plant-pathologists rely on labor-intensive and time-consuming identification of Soybean Cyst Nematode (SCN) eggs in soil samples via manual microscopy. The proposed framework attempts to significantly expedite the process by using a series of manually labeled microscopic images for training followed by automated high-throughput egg detection. The problem is particularly difficult due to the presence of a large population of non-egg particles (disturbances) in the image frames that are very similar to SCN eggs in shape, pose and illumination. Therefore, the selective autoencoder is trained to learn unique features related to the invariant shapes and sizes of the SCN eggs without handcrafting. After that, a composite non-maximum suppression and differencing is applied at the post-processing stage.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge