Arun G. Chandrasekhar
Model-Based Inference and Experimental Design for Interference Using Partial Network Data
Jun 17, 2024Abstract:The stable unit treatment value assumption states that the outcome of an individual is not affected by the treatment statuses of others, however in many real world applications, treatments can have an effect on many others beyond the immediately treated. Interference can generically be thought of as mediated through some network structure. In many empirically relevant situations however, complete network data (required to adjust for these spillover effects) are too costly or logistically infeasible to collect. Partially or indirectly observed network data (e.g., subsamples, aggregated relational data (ARD), egocentric sampling, or respondent-driven sampling) reduce the logistical and financial burden of collecting network data, but the statistical properties of treatment effect adjustments from these design strategies are only beginning to be explored. In this paper, we present a framework for the estimation and inference of treatment effect adjustments using partial network data through the lens of structural causal models. We also illustrate procedures to assign treatments using only partial network data, with the goal of either minimizing estimator variance or optimally seeding. We derive single network asymptotic results applicable to a variety of choices for an underlying graph model. We validate our approach using simulated experiments on observed graphs with applications to information diffusion in India and Malawi.
Robustly estimating heterogeneity in factorial data using Rashomon Partitions
Apr 02, 2024



Abstract:Many statistical analyses, in both observational data and randomized control trials, ask: how does the outcome of interest vary with combinations of observable covariates? How do various drug combinations affect health outcomes, or how does technology adoption depend on incentives and demographics? Our goal is to partition this factorial space into ``pools'' of covariate combinations where the outcome differs across the pools (but not within a pool). Existing approaches (i) search for a single ``optimal'' partition under assumptions about the association between covariates or (ii) sample from the entire set of possible partitions. Both these approaches ignore the reality that, especially with correlation structure in covariates, many ways to partition the covariate space may be statistically indistinguishable, despite very different implications for policy or science. We develop an alternative perspective, called Rashomon Partition Sets (RPSs). Each item in the RPS partitions the space of covariates using a tree-like geometry. RPSs incorporate all partitions that have posterior values near the maximum a posteriori partition, even if they offer substantively different explanations, and do so using a prior that makes no assumptions about associations between covariates. This prior is the $\ell_0$ prior, which we show is minimax optimal. Given the RPS we calculate the posterior of any measurable function of the feature effects vector on outcomes, conditional on being in the RPS. We also characterize approximation error relative to the entire posterior and provide bounds on the size of the RPS. Simulations demonstrate this framework allows for robust conclusions relative to conventional regularization techniques. We apply our method to three empirical settings: price effects on charitable giving, chromosomal structure (telomere length), and the introduction of microfinance.
Identifying the latent space geometry of network models through analysis of curvature
Jan 05, 2021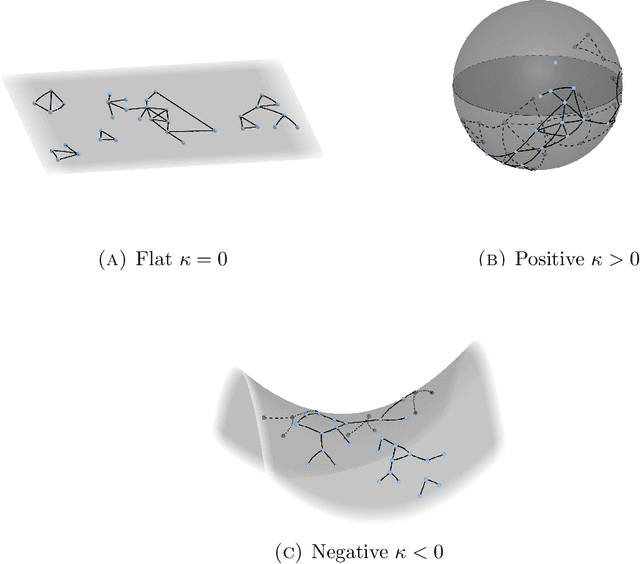

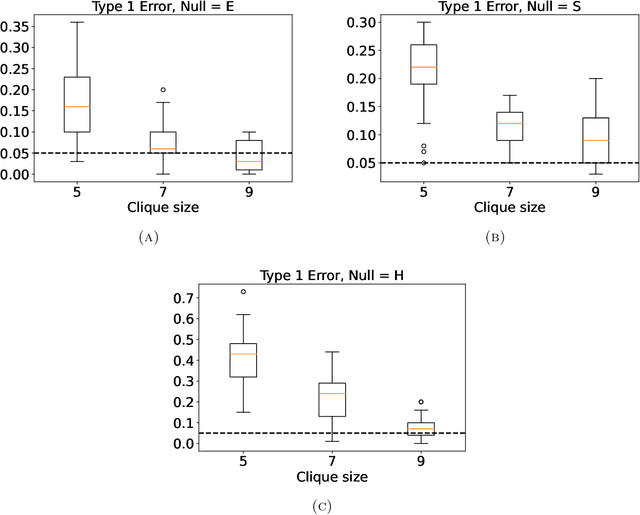

Abstract:Statistically modeling networks, across numerous disciplines and contexts, is fundamentally challenging because of (often high-order) dependence between connections. A common approach assigns each person in the graph to a position on a low-dimensional manifold. Distance between individuals in this (latent) space is inversely proportional to the likelihood of forming a connection. The choice of the latent geometry (the manifold class, dimension, and curvature) has consequential impacts on the substantive conclusions of the model. More positive curvature in the manifold, for example, encourages more and tighter communities; negative curvature induces repulsion among nodes. Currently, however, the choice of the latent geometry is an a priori modeling assumption and there is limited guidance about how to make these choices in a data-driven way. In this work, we present a method to consistently estimate the manifold type, dimension, and curvature from an empirically relevant class of latent spaces: simply connected, complete Riemannian manifolds of constant curvature. Our core insight comes by representing the graph as a noisy distance matrix based on the ties between cliques. Leveraging results from statistical geometry, we develop hypothesis tests to determine whether the observed distances could plausibly be embedded isometrically in each of the candidate geometries. We explore the accuracy of our approach with simulations and then apply our approach to data-sets from economics and sociology as well as neuroscience.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge