Anindya Sen
A Low-Cost UAV Deep Learning Pipeline for Integrated Apple Disease Diagnosis,Freshness Assessment, and Fruit Detection
Dec 28, 2025Abstract:Apple orchards require timely disease detection, fruit quality assessment, and yield estimation, yet existing UAV-based systems address such tasks in isolation and often rely on costly multispectral sensors. This paper presents a unified, low-cost RGB-only UAV-based orchard intelligent pipeline integrating ResNet50 for leaf disease detection, VGG 16 for apple freshness determination, and YOLOv8 for real-time apple detection and localization. The system runs on an ESP32-CAM and Raspberry Pi, providing fully offline on-site inference without cloud support. Experiments demonstrate 98.9% accuracy for leaf disease classification, 97.4% accuracy for freshness classification, and 0.857 F1 score for apple detection. The framework provides an accessible and scalable alternative to multispectral UAV solutions, supporting practical precision agriculture on affordable hardware.
A Comparative Study of Multiple Deep Learning Algorithms for Efficient Localization of Bone Joints in the Upper Limbs of Human Body
Oct 28, 2024Abstract:This paper addresses the medical imaging problem of joint detection in the upper limbs, viz. elbow, shoulder, wrist and finger joints. Localization of joints from X-Ray and Computerized Tomography (CT) scans is an essential step for the assessment of various bone-related medical conditions like Osteoarthritis, Rheumatoid Arthritis, and can even be used for automated bone fracture detection. Automated joint localization also detects the corresponding bones and can serve as input to deep learning-based models used for the computerized diagnosis of the aforementioned medical disorders. This in-creases the accuracy of prediction and aids the radiologists with analyzing the scans, which is quite a complex and exhausting task. This paper provides a detailed comparative study between diverse Deep Learning (DL) models - YOLOv3, YOLOv7, EfficientDet and CenterNet in multiple bone joint detections in the upper limbs of the human body. The research analyses the performance of different DL models, mathematically, graphically and visually. These models are trained and tested on a portion of the openly available MURA (musculoskeletal radiographs) dataset. The study found that the best Mean Average Precision (mAP at 0.5:0.95) values of YOLOv3, YOLOv7, EfficientDet and CenterNet are 35.3, 48.3, 46.5 and 45.9 respectively. Besides, it has been found YOLOv7 performed the best for accurately predicting the bounding boxes while YOLOv3 performed the worst in the Visual Analysis test. Code available at https://github.com/Sohambasu07/BoneJointsLocalization
A Novel Approach to Breast Cancer Histopathological Image Classification Using Cross-Colour Space Feature Fusion and Quantum-Classical Stack Ensemble Method
Apr 03, 2024Abstract:Breast cancer classification stands as a pivotal pillar in ensuring timely diagnosis and effective treatment. This study with histopathological images underscores the profound significance of harnessing the synergistic capabilities of colour space ensembling and quantum-classical stacking to elevate the precision of breast cancer classification. By delving into the distinct colour spaces of RGB, HSV and CIE L*u*v, the authors initiated a comprehensive investigation guided by advanced methodologies. Employing the DenseNet121 architecture for feature extraction the authors have capitalized on the robustness of Random Forest, SVM, QSVC, and VQC classifiers. This research encompasses a unique feature fusion technique within the colour space ensemble. This approach not only deepens our comprehension of breast cancer classification but also marks a milestone in personalized medical assessment. The amalgamation of quantum and classical classifiers through stacking emerges as a potent catalyst, effectively mitigating the inherent constraints of individual classifiers, paving a robust path towards more dependable and refined breast cancer identification. Through rigorous experimentation and meticulous analysis, fusion of colour spaces like RGB with HSV and RGB with CIE L*u*v, presents an classification accuracy, nearing the value of unity. This underscores the transformative potential of our approach, where the fusion of diverse colour spaces and the synergy of quantum and classical realms converge to establish a new horizon in medical diagnostics. Thus the implications of this research extend across medical disciplines, offering promising avenues for advancing diagnostic accuracy and treatment efficacy.
Longitudinal Volumetric Study for the Progression of Alzheimer's Disease from Structural MR Images
Oct 09, 2023Abstract:Alzheimer's Disease (AD) is primarily an irreversible neurodegenerative disorder affecting millions of individuals today. The prognosis of the disease solely depends on treating symptoms as they arise and proper caregiving, as there are no current medical preventative treatments. For this purpose, early detection of the disease at its most premature state is of paramount importance. This work aims to survey imaging biomarkers corresponding to the progression of Alzheimer's Disease (AD). A longitudinal study of structural MR images was performed for given temporal test subjects selected randomly from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database. The pipeline implemented includes modern pre-processing techniques such as spatial image registration, skull stripping, and inhomogeneity correction. The temporal data across multiple visits spanning several years helped identify the structural change in the form of volumes of cerebrospinal fluid (CSF), grey matter (GM), and white matter (WM) as the patients progressed further into the disease. Tissue classes are segmented using an unsupervised learning approach using intensity histogram information. The segmented features thus extracted provide insights such as atrophy, increase or intolerable shifting of GM, WM and CSF and should help in future research for automated analysis of Alzheimer's detection with clinical domain explainability.
Segmentation of Blood Vessels, Optic Disc Localization, Detection of Exudates and Diabetic Retinopathy Diagnosis from Digital Fundus Images
Jul 09, 2022Abstract:Diabetic Retinopathy (DR) is a complication of long-standing, unchecked diabetes and one of the leading causes of blindness in the world. This paper focuses on improved and robust methods to extract some of the features of DR, viz. Blood Vessels and Exudates. Blood vessels are segmented using multiple morphological and thresholding operations. For the segmentation of exudates, k-means clustering and contour detection on the original images are used. Extensive noise reduction is performed to remove false positives from the vessel segmentation algorithm's results. The localization of Optic Disc using k-means clustering and template matching is also performed. Lastly, this paper presents a Deep Convolutional Neural Network (DCNN) model with 14 Convolutional Layers and 2 Fully Connected Layers, for the automatic, binary diagnosis of DR. The vessel segmentation, optic disc localization and DCNN achieve accuracies of 95.93%, 98.77% and 75.73% respectively. The source code and pre-trained model are available https://github.com/Sohambasu07/DR_2021
* RAAI 2020, 11 pages, 12 figures, 2 tables
Deep Learning based Framework for Automatic Diagnosis of Glaucoma based on analysis of Focal Notching in the Optic Nerve Head
Dec 10, 2021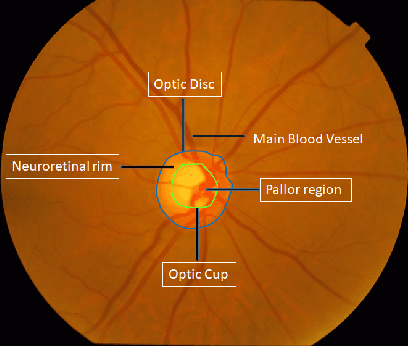
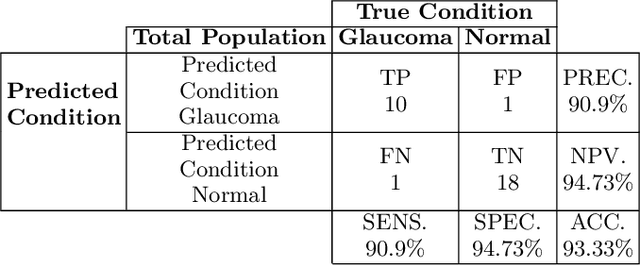
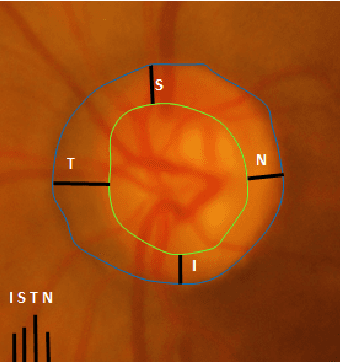
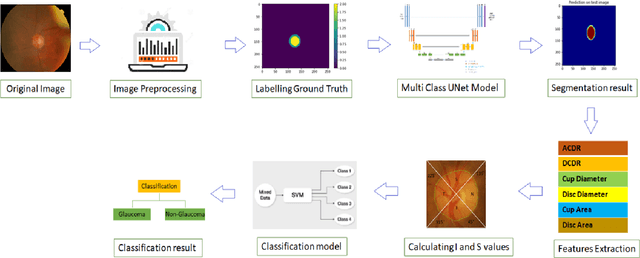
Abstract:Automatic evaluation of the retinal fundus image is emerging as one of the most important tools for early detection and treatment of progressive eye diseases like Glaucoma. Glaucoma results to a progressive degeneration of vision and is characterized by the deformation of the shape of optic cup and the degeneration of the blood vessels resulting in the formation of a notch along the neuroretinal rim. In this paper, we propose a deep learning-based pipeline for automatic segmentation of optic disc (OD) and optic cup (OC) regions from Digital Fundus Images (DFIs), thereby extracting distinct features necessary for prediction of Glaucoma. This methodology has utilized focal notch analysis of neuroretinal rim along with cup-to-disc ratio values as classifying parameters to enhance the accuracy of Computer-aided design (CAD) systems in analyzing glaucoma. Support Vector-based Machine Learning algorithm is used for classification, which classifies DFIs as Glaucomatous or Normal based on the extracted features. The proposed pipeline was evaluated on the freely available DRISHTI-GS dataset with a resultant accuracy of 93.33% for detecting Glaucoma from DFIs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge