Andrew D White
ChemCrow: Augmenting large-language models with chemistry tools
Apr 12, 2023Abstract:Large-language models (LLMs) have recently shown strong performance in tasks across domains, but struggle with chemistry-related problems. Moreover, these models lack access to external knowledge sources, limiting their usefulness in scientific applications. In this study, we introduce ChemCrow, an LLM chemistry agent designed to accomplish tasks across organic synthesis, drug discovery, and materials design. By integrating 13 expert-designed tools, ChemCrow augments the LLM performance in chemistry, and new capabilities emerge. Our evaluation, including both LLM and expert human assessments, demonstrates ChemCrow's effectiveness in automating a diverse set of chemical tasks. Surprisingly, we find that GPT-4 as an evaluator cannot distinguish between clearly wrong GPT-4 completions and GPT-4 + ChemCrow performance. There is a significant risk of misuse of tools like ChemCrow and we discuss their potential harms. Employed responsibly, ChemCrow not only aids expert chemists and lowers barriers for non-experts, but also fosters scientific advancement by bridging the gap between experimental and computational chemistry.
Classifying Antimicrobial and Multifunctional Peptides with Bayesian Network Models
Apr 17, 2018
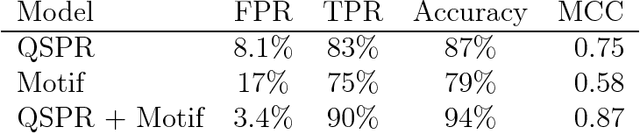


Abstract:Bayesian network models are finding success in characterizing enzyme-catalyzed reactions, slow conformational changes, predicting enzyme inhibition, and genomics. In this work, we apply them to statistical modeling of peptides by simultaneously identifying amino acid sequence motifs and using a motif-based model to clarify the role motifs may play in antimicrobial activity. We construct models of increasing sophistication, demonstrating how chemical knowledge of a peptide system may be embedded without requiring new derivation of model fitting equations after changing model structure. These models are used to construct classifiers with good performance (94% accuracy, Matthews correlation coefficient of 0.87) at predicting antimicrobial activity in peptides, while at the same time being built of interpretable parameters. We demonstrate use of these models to identify peptides that are potentially both antimicrobial and antifouling, and show that the background distribution of amino acids could play a greater role in activity than sequence motifs do. This provides an advancement in the type of peptide activity modeling that can be done and the ease in which models can be constructed.
* 19 pages, 7 figures, 1 table, supporting information included
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge