Andres Torrubia
AI Cancer Research Unit Medbravo
XDEEP-MSI: Explainable Bias-Rejecting Microsatellite Instability Deep Learning System In Colorectal Cancer
Oct 28, 2021



Abstract:We present a system for the prediction of microsatellite instability (MSI) from H&E images of colorectal cancer using deep learning (DL) techniques customized for tissue microarrays (TMAs). The system incorporates an end-to-end image preprocessing module that produces tiles at multiple magnifications in the regions of interest as guided by a tissue classifier module, and a multiple-bias rejecting module. The training and validation TMA samples were obtained from the EPICOLON project and further enriched with samples from a single institution. A systematic study of biases at tile level identified three protected (bias) variables associated with the learned representations of a baseline model: the project of origin of samples, the patient spot and the TMA glass where each spot was placed. A multiple bias rejecting technique based on adversarial training is implemented at the DL architecture so to directly avoid learning the batch effects of those variables. The learned features from the bias-ablated model have maximum discriminative power with respect to the task and minimal statistical mean dependence with the biases. The impact of different magnifications, types of tissues and the model performance at tile vs patient level is analyzed. The AUC at tile level, and including all three selected tissues (tumor epithelium, mucine and lymphocytic regions) and 4 magnifications, was 0.87 +/- 0.03 and increased to 0.9 +/- 0.03 at patient level. To the best of our knowledge, this is the first work that incorporates a multiple bias ablation technique at the DL architecture in digital pathology, and the first using TMAs for the MSI prediction task.
A community-powered search of machine learning strategy space to find NMR property prediction models
Aug 13, 2020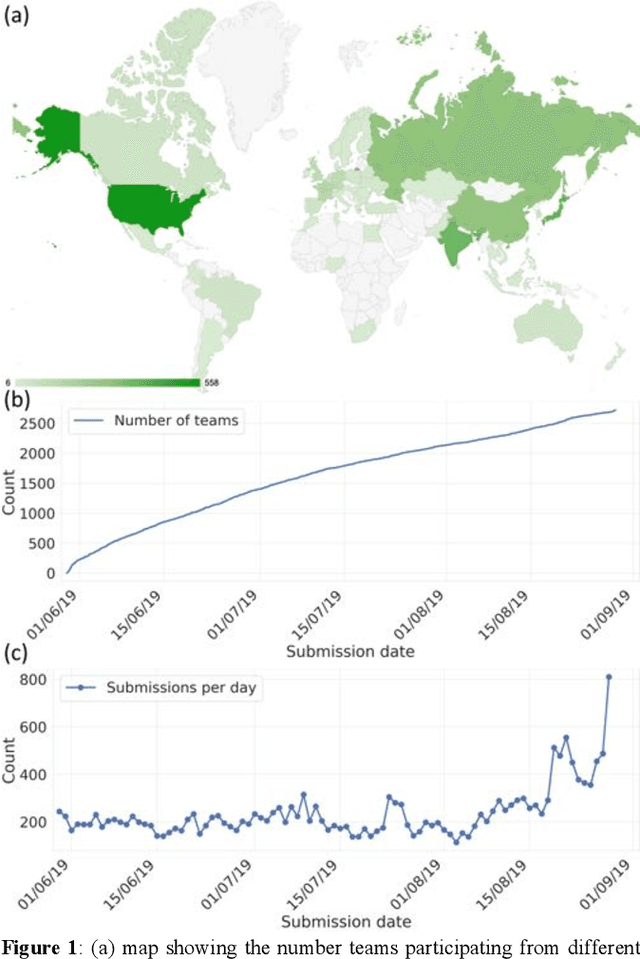
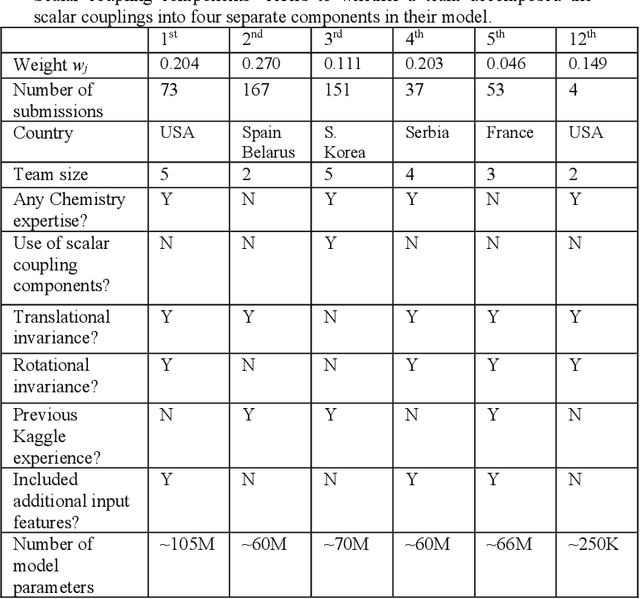
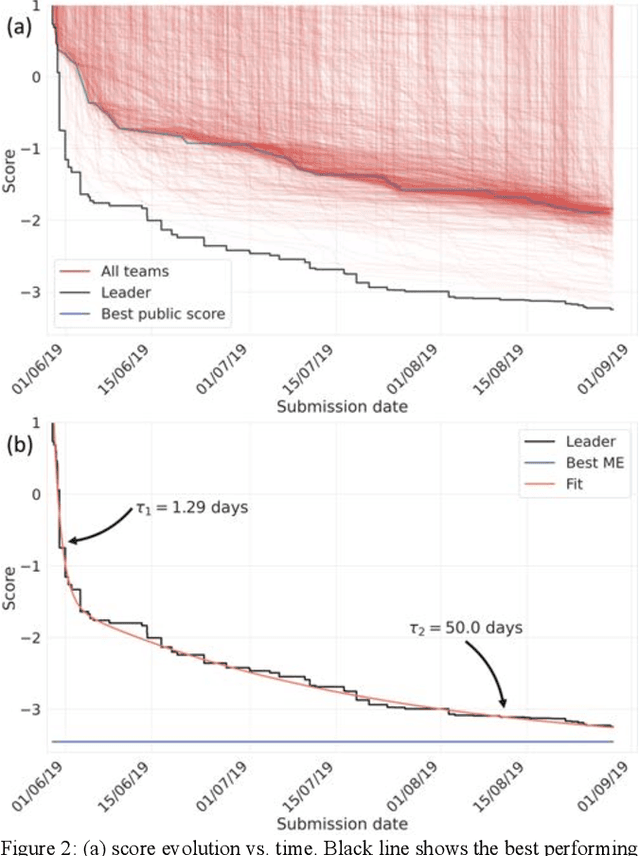
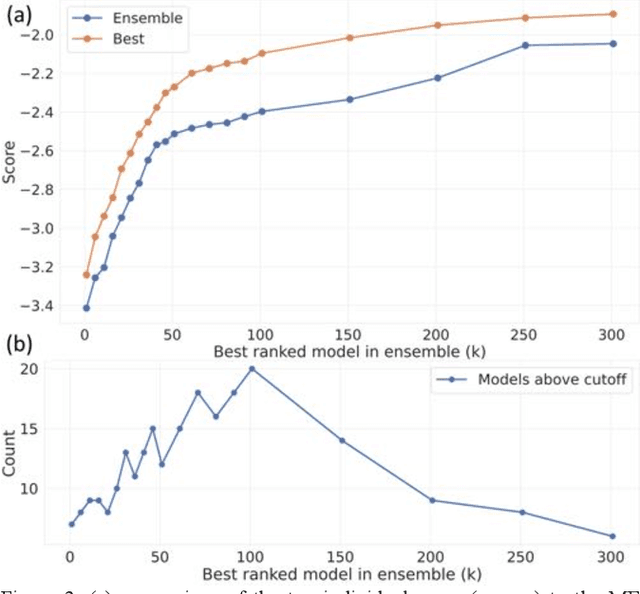
Abstract:The rise of machine learning (ML) has created an explosion in the potential strategies for using data to make scientific predictions. For physical scientists wishing to apply ML strategies to a particular domain, it can be difficult to assess in advance what strategy to adopt within a vast space of possibilities. Here we outline the results of an online community-powered effort to swarm search the space of ML strategies and develop algorithms for predicting atomic-pairwise nuclear magnetic resonance (NMR) properties in molecules. Using an open-source dataset, we worked with Kaggle to design and host a 3-month competition which received 47,800 ML model predictions from 2,700 teams in 84 countries. Within 3 weeks, the Kaggle community produced models with comparable accuracy to our best previously published "in-house" efforts. A meta-ensemble model constructed as a linear combination of the top predictions has a prediction accuracy which exceeds that of any individual model, 7-19x better than our previous state-of-the-art. The results highlight the potential of transformer architectures for predicting quantum mechanical (QM) molecular properties.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge