Alireza Moradzadeh
BioNeMo Framework: a modular, high-performance library for AI model development in drug discovery
Nov 15, 2024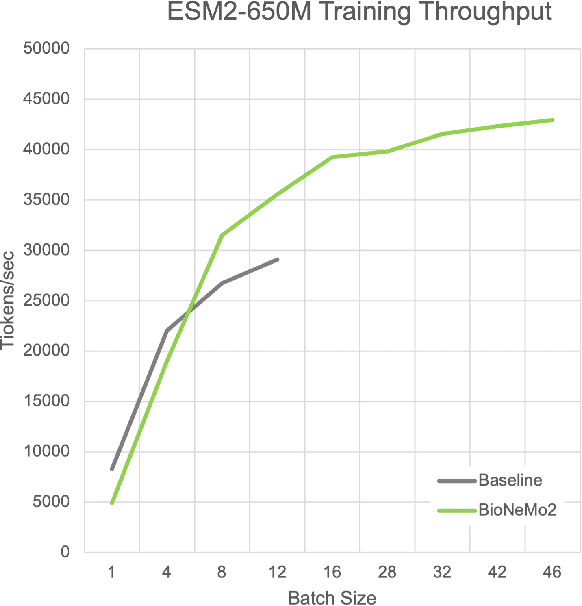
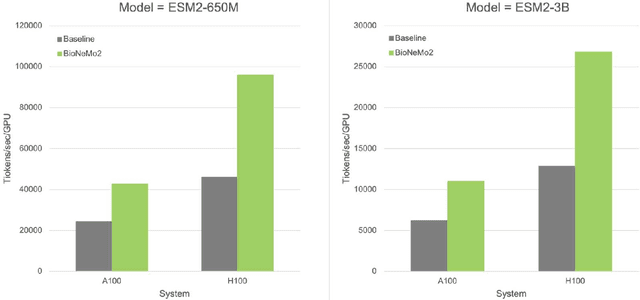
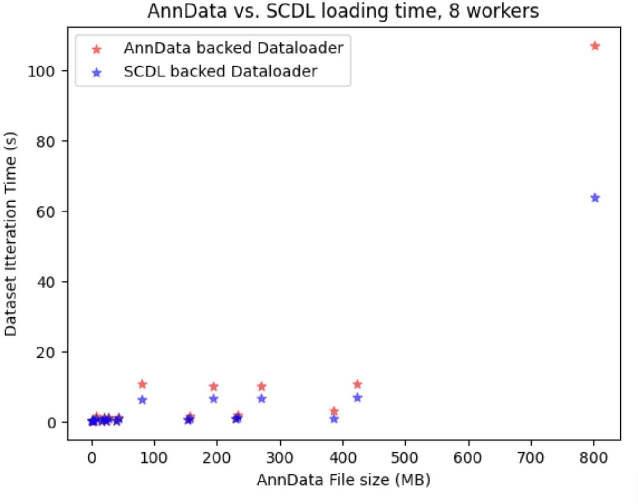
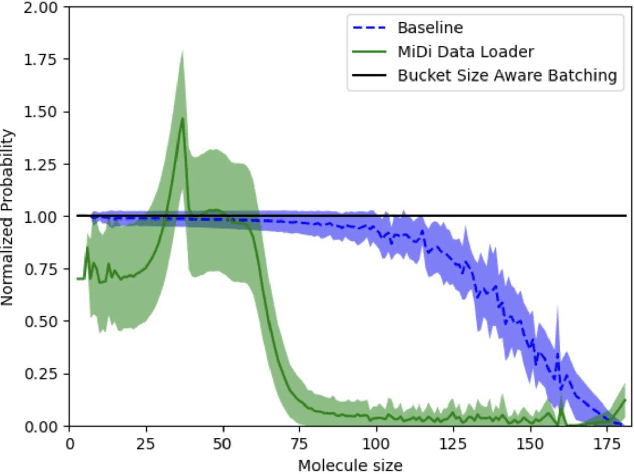
Abstract:Artificial Intelligence models encoding biology and chemistry are opening new routes to high-throughput and high-quality in-silico drug development. However, their training increasingly relies on computational scale, with recent protein language models (pLM) training on hundreds of graphical processing units (GPUs). We introduce the BioNeMo Framework to facilitate the training of computational biology and chemistry AI models across hundreds of GPUs. Its modular design allows the integration of individual components, such as data loaders, into existing workflows and is open to community contributions. We detail technical features of the BioNeMo Framework through use cases such as pLM pre-training and fine-tuning. On 256 NVIDIA A100s, BioNeMo Framework trains a three billion parameter BERT-based pLM on over one trillion tokens in 4.2 days. The BioNeMo Framework is open-source and free for everyone to use.
UKAN: Unbound Kolmogorov-Arnold Network Accompanied with Accelerated Library
Aug 20, 2024



Abstract:In this work, we present a GPU-accelerated library for the underlying components of Kolmogorov-Arnold Networks (KANs), along with an algorithm to eliminate bounded grids in KANs. The GPU-accelerated library reduces the computational complexity of Basis Spline (B-spline) evaluation by a factor of $\mathcal{O}$(grid size) compared to existing codes, enabling batch computation for large-scale learning. To overcome the limitations of traditional KANs, we introduce Unbounded KANs (UKANs), which eliminate the need for a bounded grid and a fixed number of B-spline coefficients. To do so, we replace the KAN parameters (B-spline coefficients) with a coefficient generator (CG) model. The inputs to the CG model are designed based on the idea of an infinite symmetric grid extending from negative infinity to positive infinity. The positional encoding of grid group, a sequential collection of B-spline grid indexes, is fed into the CG model, and coefficients are consumed by the efficient implementation (matrix representations) of B-spline functions to generate outputs. We perform several experiments on regression, classification, and generative tasks, which are promising. In particular, UKAN does not require data normalization or a bounded domain for evaluation. Additionally, our benchmarking results indicate the superior memory and computational efficiency of our library compared to existing codes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge