Alexander S. Yeh
Mitre Corporation
Evaluation of text data mining for database curation: lessons learned from the KDD Challenge Cup
Aug 20, 2003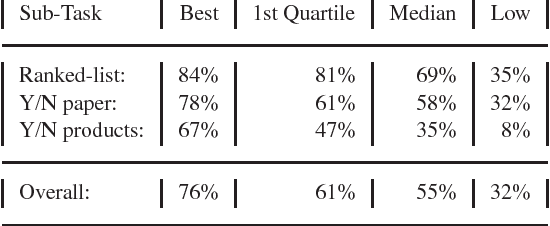
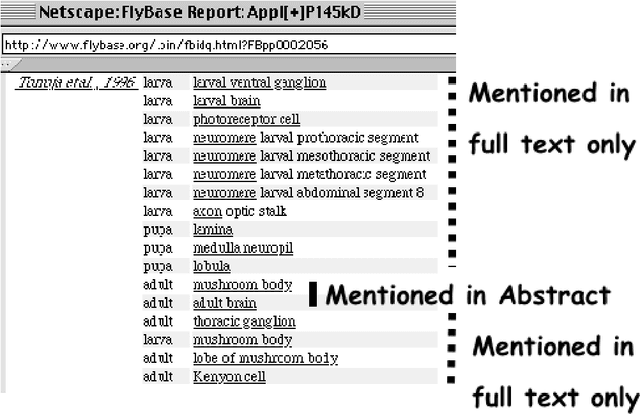

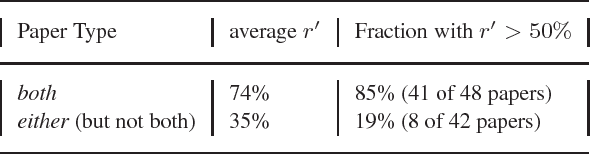
Abstract:MOTIVATION: The biological literature is a major repository of knowledge. Many biological databases draw much of their content from a careful curation of this literature. However, as the volume of literature increases, the burden of curation increases. Text mining may provide useful tools to assist in the curation process. To date, the lack of standards has made it impossible to determine whether text mining techniques are sufficiently mature to be useful. RESULTS: We report on a Challenge Evaluation task that we created for the Knowledge Discovery and Data Mining (KDD) Challenge Cup. We provided a training corpus of 862 articles consisting of journal articles curated in FlyBase, along with the associated lists of genes and gene products, as well as the relevant data fields from FlyBase. For the test, we provided a corpus of 213 new (`blind') articles; the 18 participating groups provided systems that flagged articles for curation, based on whether the article contained experimental evidence for gene expression products. We report on the the evaluation results and describe the techniques used by the top performing groups. CONTACT: asy@mitre.org KEYWORDS: text mining, evaluation, curation, genomics, data management
* 9 pages. This is close to how it appears on the publisher's website (http://bioinformatics.oupjournals.org/cgi/reprint/19/suppl_1/i331) The article wording is the same. Uses bioinformatics-altered.cls, bioinformaticsbib.sty, bioinformaticstitle.sty
Some Properties of Preposition and Subordinate Conjunction Attachments
Aug 20, 1998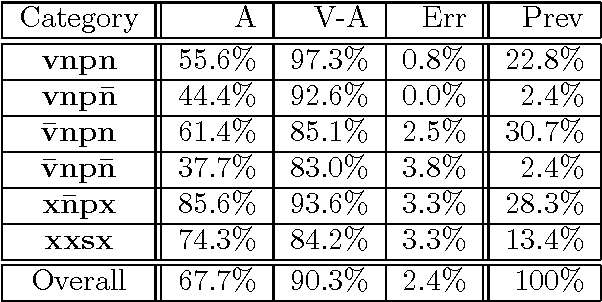
Abstract:Determining the attachments of prepositions and subordinate conjunctions is a key problem in parsing natural language. This paper presents a trainable approach to making these attachments through transformation sequences and error-driven learning. Our approach is broad coverage, and accounts for roughly three times the attachment cases that have previously been handled by corpus-based techniques. In addition, our approach is based on a simplified model of syntax that is more consistent with the practice in current state-of-the-art language processing systems. This paper sketches syntactic and algorithmic details, and presents experimental results on data sets derived from the Penn Treebank. We obtain an attachment accuracy of 75.4% for the general case, the first such corpus-based result to be reported. For the restricted cases previously studied with corpus-based methods, our approach yields an accuracy comparable to current work (83.1%).
* 7 pages, uses colacl.sty
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge