Alex A. T. Bui
on behalf of CURE-CKD
Bridge2AI: Building A Cross-disciplinary Curriculum Towards AI-Enhanced Biomedical and Clinical Care
May 20, 2025Abstract:Objective: As AI becomes increasingly central to healthcare, there is a pressing need for bioinformatics and biomedical training systems that are personalized and adaptable. Materials and Methods: The NIH Bridge2AI Training, Recruitment, and Mentoring (TRM) Working Group developed a cross-disciplinary curriculum grounded in collaborative innovation, ethical data stewardship, and professional development within an adapted Learning Health System (LHS) framework. Results: The curriculum integrates foundational AI modules, real-world projects, and a structured mentee-mentor network spanning Bridge2AI Grand Challenges and the Bridge Center. Guided by six learner personas, the program tailors educational pathways to individual needs while supporting scalability. Discussion: Iterative refinement driven by continuous feedback ensures that content remains responsive to learner progress and emerging trends. Conclusion: With over 30 scholars and 100 mentors engaged across North America, the TRM model demonstrates how adaptive, persona-informed training can build interdisciplinary competencies and foster an integrative, ethically grounded AI education in biomedical contexts.
AKIBoards: A Structure-Following Multiagent System for Predicting Acute Kidney Injury
Apr 29, 2025Abstract:Diagnostic reasoning entails a physician's local (mental) model based on an assumed or known shared perspective (global model) to explain patient observations with evidence assigned towards a clinical assessment. But in several (complex) medical situations, multiple experts work together as a team to optimize health evaluation and decision-making by leveraging different perspectives. Such consensus-driven reasoning reflects individual knowledge contributing toward a broader perspective on the patient. In this light, we introduce STRUCture-following for Multiagent Systems (STRUC-MAS), a framework automating the learning of these global models and their incorporation as prior beliefs for agents in multiagent systems (MAS) to follow. We demonstrate proof of concept with a prosocial MAS application for predicting acute kidney injuries (AKIs). In this case, we found that incorporating a global structure enabled multiple agents to achieve better performance (average precision, AP) in predicting AKI 48 hours before onset (structure-following-fine-tuned, SF-FT, AP=0.195; SF-FT-retrieval-augmented generation, SF-FT-RAG, AP=0.194) vs. baseline (non-structure-following-FT, NSF-FT, AP=0.141; NSF-FT-RAG, AP=0.180) for balanced precision-weighted-recall-weighted voting. Markedly, SF-FT agents with higher recall scores reported lower confidence levels in the initial round on true positive and false negative cases. But after explicit interactions, their confidence in their decisions increased (suggesting reinforced belief). In contrast, the SF-FT agent with the lowest recall decreased its confidence in true positive and false negative cases (suggesting a new belief). This approach suggests that learning and leveraging global structures in MAS is necessary prior to achieving competitive classification and diagnostic reasoning performance.
Testing Causal Explanations: A Case Study for Understanding the Effect of Interventions on Chronic Kidney Disease
Oct 15, 2024Abstract:Randomized controlled trials (RCTs) are the standard for evaluating the effectiveness of clinical interventions. To address the limitations of RCTs on real-world populations, we developed a methodology that uses a large observational electronic health record (EHR) dataset. Principles of regression discontinuity (rd) were used to derive randomized data subsets to test expert-driven interventions using dynamic Bayesian Networks (DBNs) do-operations. This combined method was applied to a chronic kidney disease (CKD) cohort of more than two million individuals and used to understand the associational and causal relationships of CKD variables with respect to a surrogate outcome of >=40% decline in estimated glomerular filtration rate (eGFR). The associational and causal analyses depicted similar findings across DBNs from two independent healthcare systems. The associational analysis showed that the most influential variables were eGFR, urine albumin-to-creatinine ratio, and pulse pressure, whereas the causal analysis showed eGFR as the most influential variable, followed by modifiable factors such as medications that may impact kidney function over time. This methodology demonstrates how real-world EHR data can be used to provide population-level insights to inform improved healthcare delivery.
Automated Dynamic Bayesian Networks for Predicting Acute Kidney Injury Before Onset
Apr 20, 2023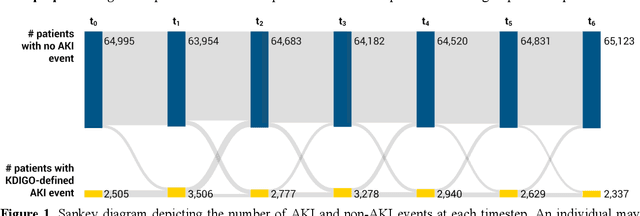



Abstract:Several algorithms for learning the structure of dynamic Bayesian networks (DBNs) require an a priori ordering of variables, which influences the determined graph topology. However, it is often unclear how to determine this order if feature importance is unknown, especially as an exhaustive search is usually impractical. In this paper, we introduce Ranking Approaches for Unknown Structures (RAUS), an automated framework to systematically inform variable ordering and learn networks end-to-end. RAUS leverages existing statistical methods (Cramers V, chi-squared test, and information gain) to compare variable ordering, resultant generated network topologies, and DBN performance. RAUS enables end-users with limited DBN expertise to implement models via command line interface. We evaluate RAUS on the task of predicting impending acute kidney injury (AKI) from inpatient clinical laboratory data. Longitudinal observations from 67,460 patients were collected from our electronic health record (EHR) and Kidney Disease Improving Global Outcomes (KDIGO) criteria were then applied to define AKI events. RAUS learns multiple DBNs simultaneously to predict a future AKI event at different time points (i.e., 24-, 48-, 72-hours in advance of AKI). We also compared the results of the learned AKI prediction models and variable orderings to baseline techniques (logistic regression, random forests, and extreme gradient boosting). The DBNs generated by RAUS achieved 73-83% area under the receiver operating characteristic curve (AUCROC) within 24-hours before AKI; and 71-79% AUCROC within 48-hours before AKI of any stage in a 7-day observation window. Insights from this automated framework can help efficiently implement and interpret DBNs for clinical decision support. The source code for RAUS is available in GitHub at https://github.com/dgrdn08/RAUS .
An Interpretable Deep Hierarchical Semantic Convolutional Neural Network for Lung Nodule Malignancy Classification
Jun 02, 2018



Abstract:While deep learning methods are increasingly being applied to tasks such as computer-aided diagnosis, these models are difficult to interpret, do not incorporate prior domain knowledge, and are often considered as a "black-box." The lack of model interpretability hinders them from being fully understood by target users such as radiologists. In this paper, we present a novel interpretable deep hierarchical semantic convolutional neural network (HSCNN) to predict whether a given pulmonary nodule observed on a computed tomography (CT) scan is malignant. Our network provides two levels of output: 1) low-level radiologist semantic features, and 2) a high-level malignancy prediction score. The low-level semantic outputs quantify the diagnostic features used by radiologists and serve to explain how the model interprets the images in an expert-driven manner. The information from these low-level tasks, along with the representations learned by the convolutional layers, are then combined and used to infer the high-level task of predicting nodule malignancy. This unified architecture is trained by optimizing a global loss function including both low- and high-level tasks, thereby learning all the parameters within a joint framework. Our experimental results using the Lung Image Database Consortium (LIDC) show that the proposed method not only produces interpretable lung cancer predictions but also achieves significantly better results compared to common 3D CNN approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge