Automated Dynamic Bayesian Networks for Predicting Acute Kidney Injury Before Onset
Paper and Code
Apr 20, 2023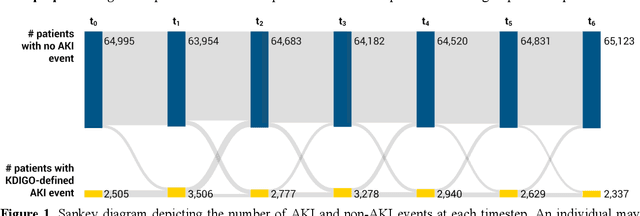



Several algorithms for learning the structure of dynamic Bayesian networks (DBNs) require an a priori ordering of variables, which influences the determined graph topology. However, it is often unclear how to determine this order if feature importance is unknown, especially as an exhaustive search is usually impractical. In this paper, we introduce Ranking Approaches for Unknown Structures (RAUS), an automated framework to systematically inform variable ordering and learn networks end-to-end. RAUS leverages existing statistical methods (Cramers V, chi-squared test, and information gain) to compare variable ordering, resultant generated network topologies, and DBN performance. RAUS enables end-users with limited DBN expertise to implement models via command line interface. We evaluate RAUS on the task of predicting impending acute kidney injury (AKI) from inpatient clinical laboratory data. Longitudinal observations from 67,460 patients were collected from our electronic health record (EHR) and Kidney Disease Improving Global Outcomes (KDIGO) criteria were then applied to define AKI events. RAUS learns multiple DBNs simultaneously to predict a future AKI event at different time points (i.e., 24-, 48-, 72-hours in advance of AKI). We also compared the results of the learned AKI prediction models and variable orderings to baseline techniques (logistic regression, random forests, and extreme gradient boosting). The DBNs generated by RAUS achieved 73-83% area under the receiver operating characteristic curve (AUCROC) within 24-hours before AKI; and 71-79% AUCROC within 48-hours before AKI of any stage in a 7-day observation window. Insights from this automated framework can help efficiently implement and interpret DBNs for clinical decision support. The source code for RAUS is available in GitHub at https://github.com/dgrdn08/RAUS .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge