Alberto Parravicini
Demystifying Drug Repurposing Domain Comprehension with Knowledge Graph Embedding
Aug 30, 2021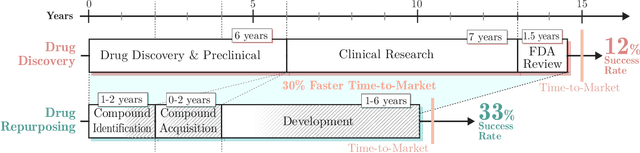
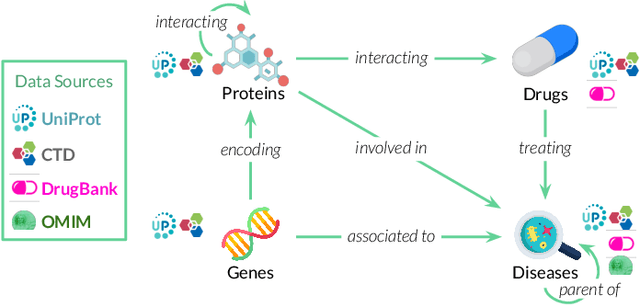
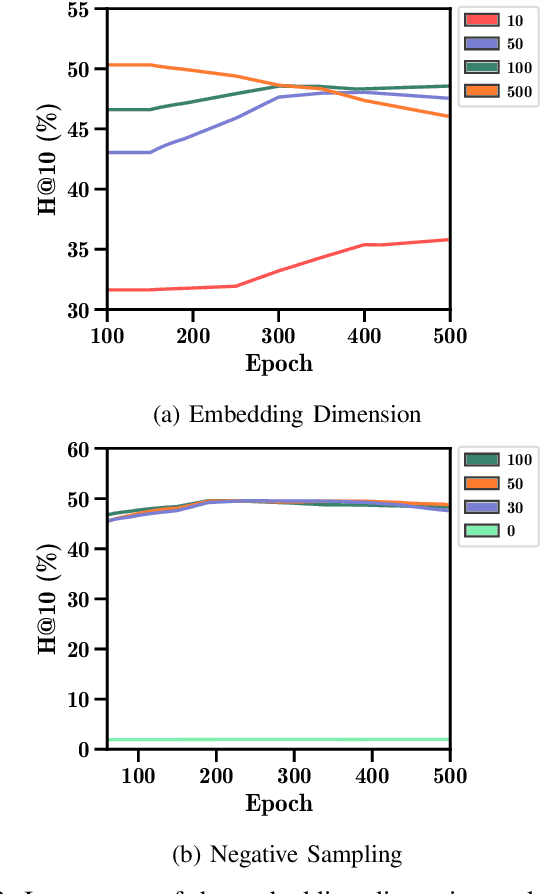
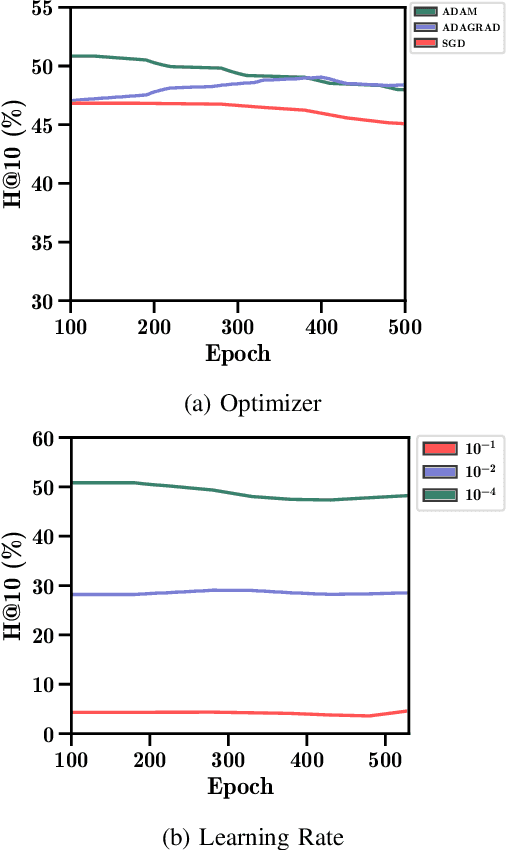
Abstract:Drug repurposing is more relevant than ever due to drug development's rising costs and the need to respond to emerging diseases quickly. Knowledge graph embedding enables drug repurposing using heterogeneous data sources combined with state-of-the-art machine learning models to predict new drug-disease links in the knowledge graph. As in many machine learning applications, significant work is still required to understand the predictive models' behavior. We propose a structured methodology to understand better machine learning models' results for drug repurposing, suggesting key elements of the knowledge graph to improve predictions while saving computational resources. We reduce the training set of 11.05% and the embedding space by 31.87%, with only a 2% accuracy reduction, and increase accuracy by 60% on the open ogbl-biokg graph adding only 1.53% new triples.
Scaling up HBM Efficiency of Top-K SpMV for Approximate Embedding Similarity on FPGAs
Mar 08, 2021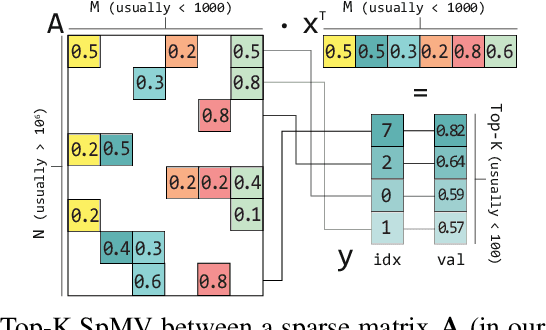
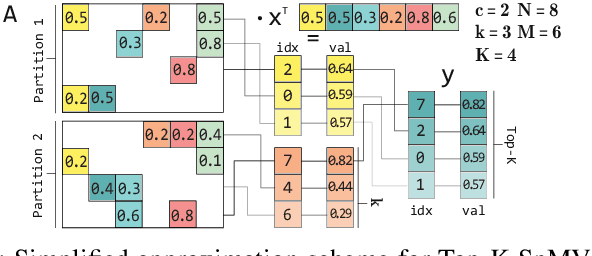
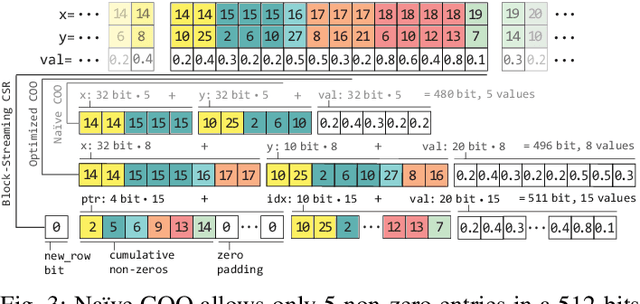
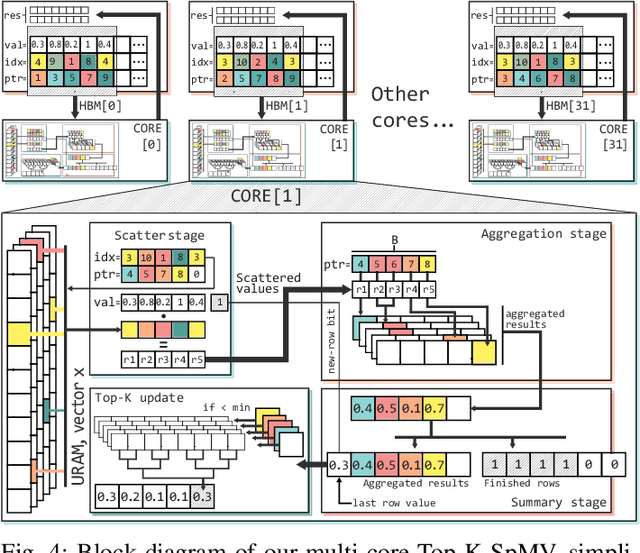
Abstract:Top-K SpMV is a key component of similarity-search on sparse embeddings. This sparse workload does not perform well on general-purpose NUMA systems that employ traditional caching strategies. Instead, modern FPGA accelerator cards have a few tricks up their sleeve. We introduce a Top-K SpMV FPGA design that leverages reduced precision and a novel packet-wise CSR matrix compression, enabling custom data layouts and delivering bandwidth efficiency often unreachable even in architectures with higher peak bandwidth. With HBM-based boards, we are 100x faster than a multi-threaded CPU implementation and 2x faster than a GPU with 20% higher bandwidth, with 14.2x higher power-efficiency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge