Alan D. Kaplan
Sequential Inference of Hospitalization ElectronicHealth Records Using Probabilistic Models
Mar 27, 2024Abstract:In the dynamic hospital setting, decision support can be a valuable tool for improving patient outcomes. Data-driven inference of future outcomes is challenging in this dynamic setting, where long sequences such as laboratory tests and medications are updated frequently. This is due in part to heterogeneity of data types and mixed-sequence types contained in variable length sequences. In this work we design a probabilistic unsupervised model for multiple arbitrary-length sequences contained in hospitalization Electronic Health Record (EHR) data. The model uses a latent variable structure and captures complex relationships between medications, diagnoses, laboratory tests, neurological assessments, and medications. It can be trained on original data, without requiring any lossy transformations or time binning. Inference algorithms are derived that use partial data to infer properties of the complete sequences, including their length and presence of specific values. We train this model on data from subjects receiving medical care in the Kaiser Permanente Northern California integrated healthcare delivery system. The results are evaluated against held-out data for predicting the length of sequences and presence of Intensive Care Unit (ICU) in hospitalization bed sequences. Our method outperforms a baseline approach, showing that in these experiments the trained model captures information in the sequences that is informative of their future values.
Unsupervised Probabilistic Models for Sequential Electronic Health Records
Apr 15, 2022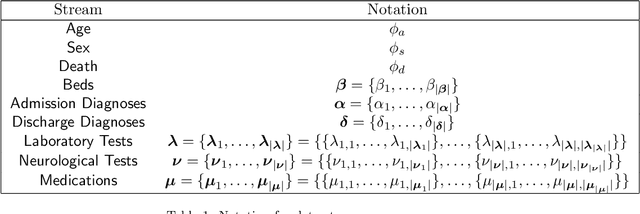
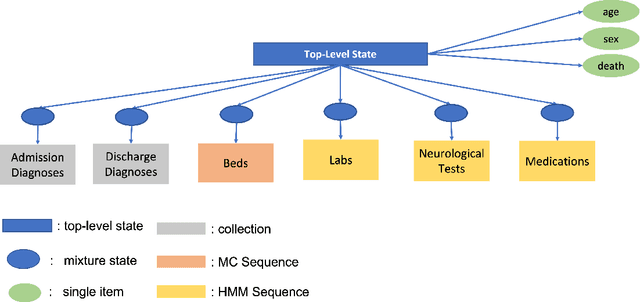
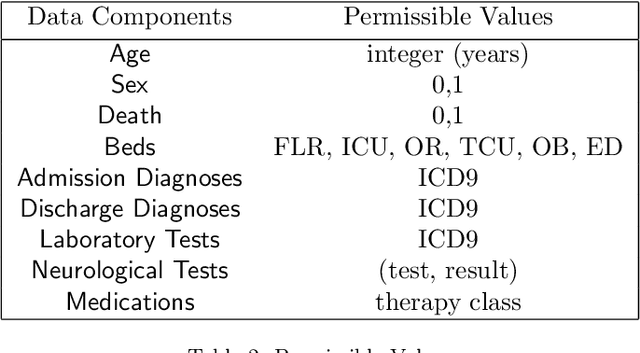
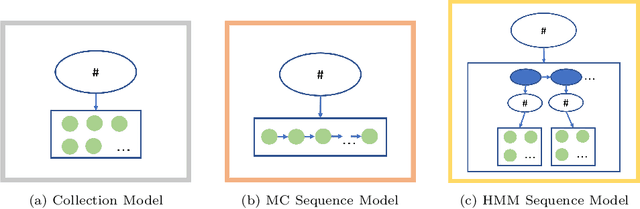
Abstract:We develop an unsupervised probabilistic model for heterogeneous Electronic Health Record (EHR) data. Utilizing a mixture model formulation, our approach directly models sequences of arbitrary length, such as medications and laboratory results. This allows for subgrouping and incorporation of the dynamics underlying heterogeneous data types. The model consists of a layered set of latent variables that encode underlying structure in the data. These variables represent subject subgroups at the top layer, and unobserved states for sequences in the second layer. We train this model on episodic data from subjects receiving medical care in the Kaiser Permanente Northern California integrated healthcare delivery system. The resulting properties of the trained model generate novel insight from these complex and multifaceted data. In addition, we show how the model can be used to analyze sequences that contribute to assessment of mortality likelihood.
Mixture Model Framework for Traumatic Brain Injury Prognosis Using Heterogeneous Clinical and Outcome Data
Dec 22, 2020



Abstract:Prognoses of Traumatic Brain Injury (TBI) outcomes are neither easily nor accurately determined from clinical indicators. This is due in part to the heterogeneity of damage inflicted to the brain, ultimately resulting in diverse and complex outcomes. Using a data-driven approach on many distinct data elements may be necessary to describe this large set of outcomes and thereby robustly depict the nuanced differences among TBI patients' recovery. In this work, we develop a method for modeling large heterogeneous data types relevant to TBI. Our approach is geared toward the probabilistic representation of mixed continuous and discrete variables with missing values. The model is trained on a dataset encompassing a variety of data types, including demographics, blood-based biomarkers, and imaging findings. In addition, it includes a set of clinical outcome assessments at 3, 6, and 12 months post-injury. The model is used to stratify patients into distinct groups in an unsupervised learning setting. We use the model to infer outcomes using input data, and show that the collection of input data reduces uncertainty of outcomes over a baseline approach. In addition, we quantify the performance of a likelihood scoring technique that can be used to self-evaluate confidence in model fit and prediction.
AutoAtlas: Neural Network for 3D Unsupervised Partitioning and Representation Learning
Nov 05, 2020



Abstract:We present a novel neural network architecture called AutoAtlas for fully unsupervised partitioning and representation learning of 3D brain Magnetic Resonance Imaging (MRI) volumes. AutoAtlas consists of two neural network components: one that performs multi-label partitioning based on local texture in the volume and a second that compresses the information contained within each partition. We train both of these components simultaneously by optimizing a loss function that is designed to promote accurate reconstruction of each partition, while encouraging spatially smooth and contiguous partitioning, and discouraging relatively small partitions. We show that the partitions adapt to the subject specific structural variations of brain tissue while consistently appearing at similar spatial locations across subjects. AutoAtlas also produces very low dimensional features that represent local texture of each partition. We demonstrate prediction of metadata associated with each subject using the derived feature representations and compare the results to prediction using features derived from FreeSurfer anatomical parcellation. Since our features are intrinsically linked to distinct partitions, we can then map values of interest, such as partition-specific feature importance scores onto the brain for visualization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge