Aiman Farooq
RobSurv: Vector Quantization-Based Multi-Modal Learning for Robust Cancer Survival Prediction
May 05, 2025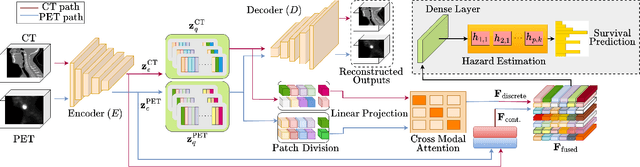
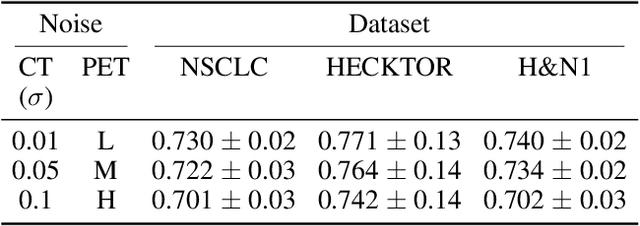
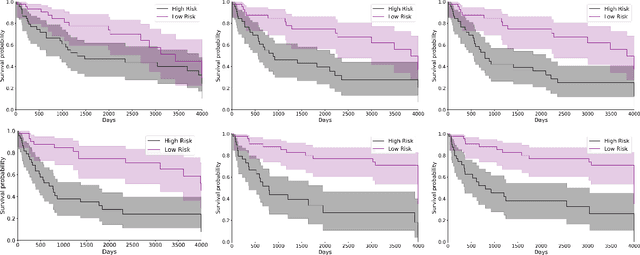
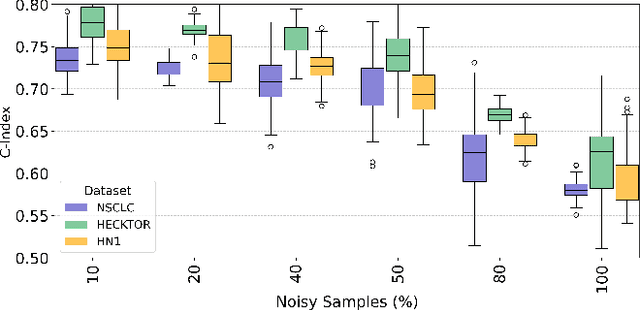
Abstract:Cancer survival prediction using multi-modal medical imaging presents a critical challenge in oncology, mainly due to the vulnerability of deep learning models to noise and protocol variations across imaging centers. Current approaches struggle to extract consistent features from heterogeneous CT and PET images, limiting their clinical applicability. We address these challenges by introducing RobSurv, a robust deep-learning framework that leverages vector quantization for resilient multi-modal feature learning. The key innovation of our approach lies in its dual-path architecture: one path maps continuous imaging features to learned discrete codebooks for noise-resistant representation, while the parallel path preserves fine-grained details through continuous feature processing. This dual representation is integrated through a novel patch-wise fusion mechanism that maintains local spatial relationships while capturing global context via Transformer-based processing. In extensive evaluations across three diverse datasets (HECKTOR, H\&N1, and NSCLC Radiogenomics), RobSurv demonstrates superior performance, achieving concordance index of 0.771, 0.742, and 0.734 respectively - significantly outperforming existing methods. Most notably, our model maintains robust performance even under severe noise conditions, with performance degradation of only 3.8-4.5\% compared to 8-12\% in baseline methods. These results, combined with strong generalization across different cancer types and imaging protocols, establish RobSurv as a promising solution for reliable clinical prognosis that can enhance treatment planning and patient care.
Fine-Grained Rib Fracture Diagnosis with Hyperbolic Embeddings: A Detailed Annotation Framework and Multi-Label Classification Model
Apr 16, 2025Abstract:Accurate rib fracture identification and classification are essential for treatment planning. However, existing datasets often lack fine-grained annotations, particularly regarding rib fracture characterization, type, and precise anatomical location on individual ribs. To address this, we introduce a novel rib fracture annotation protocol tailored for fracture classification. Further, we enhance fracture classification by leveraging cross-modal embeddings that bridge radiological images and clinical descriptions. Our approach employs hyperbolic embeddings to capture the hierarchical nature of fracture, mapping visual features and textual descriptions into a shared non-Euclidean manifold. This framework enables more nuanced similarity computations between imaging characteristics and clinical descriptions, accounting for the inherent hierarchical relationships in fracture taxonomy. Experimental results demonstrate that our approach outperforms existing methods across multiple classification tasks, with average recall improvements of 6% on the AirRib dataset and 17.5% on the public RibFrac dataset.
RibCageImp: A Deep Learning Framework for 3D Ribcage Implant Generation
Nov 14, 2024Abstract:The recovery of damaged or resected ribcage structures requires precise, custom-designed implants to restore the integrity and functionality of the thoracic cavity. Traditional implant design methods rely mainly on manual processes, making them time-consuming and susceptible to variability. In this work, we explore the feasibility of automated ribcage implant generation using deep learning. We present a framework based on 3D U-Net architecture that processes CT scans to generate patient-specific implant designs. To the best of our knowledge, this is the first investigation into automated thoracic implant generation using deep learning approaches. Our preliminary results, while moderate, highlight both the potential and the significant challenges in this complex domain. These findings establish a foundation for future research in automated ribcage reconstruction and identify key technical challenges that need to be addressed for practical implementation.
Leveraging Auxiliary Classification for Rib Fracture Segmentation
Nov 14, 2024



Abstract:Thoracic trauma often results in rib fractures, which demand swift and accurate diagnosis for effective treatment. However, detecting these fractures on rib CT scans poses considerable challenges, involving the analysis of many image slices in sequence. Despite notable advancements in algorithms for automated fracture segmentation, the persisting challenges stem from the diverse shapes and sizes of these fractures. To address these issues, this study introduces a sophisticated deep-learning model with an auxiliary classification task designed to enhance the accuracy of rib fracture segmentation. The auxiliary classification task is crucial in distinguishing between fractured ribs and negative regions, encompassing non-fractured ribs and surrounding tissues, from the patches obtained from CT scans. By leveraging this auxiliary task, the model aims to improve feature representation at the bottleneck layer by highlighting the regions of interest. Experimental results on the RibFrac dataset demonstrate significant improvement in segmentation performance.
Enhanced Survival Prediction in Head and Neck Cancer Using Convolutional Block Attention and Multimodal Data Fusion
Oct 29, 2024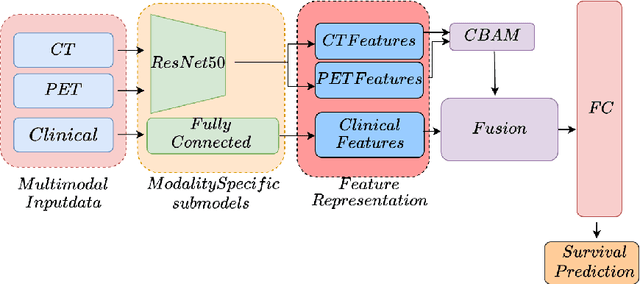
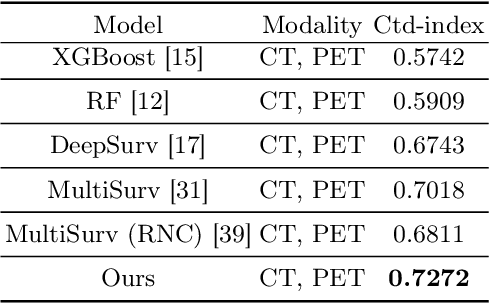
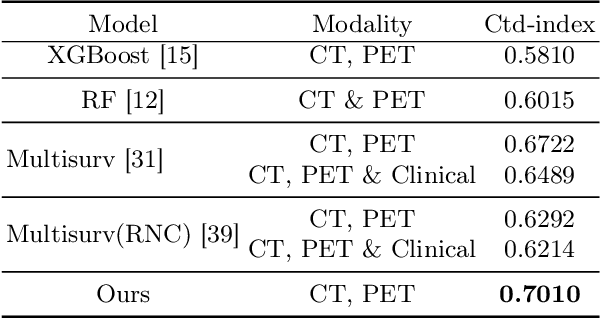
Abstract:Accurate survival prediction in head and neck cancer (HNC) is essential for guiding clinical decision-making and optimizing treatment strategies. Traditional models, such as Cox proportional hazards, have been widely used but are limited in their ability to handle complex multi-modal data. This paper proposes a deep learning-based approach leveraging CT and PET imaging modalities to predict survival outcomes in HNC patients. Our method integrates feature extraction with a Convolutional Block Attention Module (CBAM) and a multi-modal data fusion layer that combines imaging data to generate a compact feature representation. The final prediction is achieved through a fully parametric discrete-time survival model, allowing for flexible hazard functions that overcome the limitations of traditional survival models. We evaluated our approach using the HECKTOR and HEAD-NECK-RADIOMICS- HN1 datasets, demonstrating its superior performance compared to conconventional statistical and machine learning models. The results indicate that our deep learning model significantly improves survival prediction accuracy, offering a robust tool for personalized treatment planning in HNC
Survival Prediction in Lung Cancer through Multi-Modal Representation Learning
Sep 30, 2024



Abstract:Survival prediction is a crucial task associated with cancer diagnosis and treatment planning. This paper presents a novel approach to survival prediction by harnessing comprehensive information from CT and PET scans, along with associated Genomic data. Current methods rely on either a single modality or the integration of multiple modalities for prediction without adequately addressing associations across patients or modalities. We aim to develop a robust predictive model for survival outcomes by integrating multi-modal imaging data with genetic information while accounting for associations across patients and modalities. We learn representations for each modality via a self-supervised module and harness the semantic similarities across the patients to ensure the embeddings are aligned closely. However, optimizing solely for global relevance is inadequate, as many pairs sharing similar high-level semantics, such as tumor type, are inadvertently pushed apart in the embedding space. To address this issue, we use a cross-patient module (CPM) designed to harness inter-subject correspondences. The CPM module aims to bring together embeddings from patients with similar disease characteristics. Our experimental evaluation of the dataset of Non-Small Cell Lung Cancer (NSCLC) patients demonstrates the effectiveness of our approach in predicting survival outcomes, outperforming state-of-the-art methods.
Translating Imaging to Genomics: Leveraging Transformers for Predictive Modeling
Aug 01, 2024Abstract:In this study, we present a novel approach for predicting genomic information from medical imaging modalities using a transformer-based model. We aim to bridge the gap between imaging and genomics data by leveraging transformer networks, allowing for accurate genomic profile predictions from CT/MRI images. Presently most studies rely on the use of whole slide images (WSI) for the association, which are obtained via invasive methodologies. We propose using only available CT/MRI images to predict genomic sequences. Our transformer based approach is able to efficiently generate associations between multiple sequences based on CT/MRI images alone. This work paves the way for the use of non-invasive imaging modalities for precise and personalized healthcare, allowing for a better understanding of diseases and treatment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge