Azad Singh
RobSurv: Vector Quantization-Based Multi-Modal Learning for Robust Cancer Survival Prediction
May 05, 2025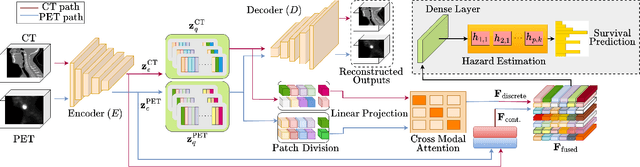
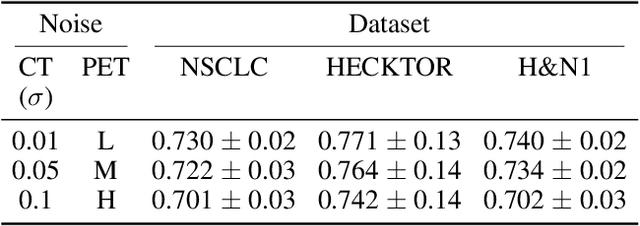
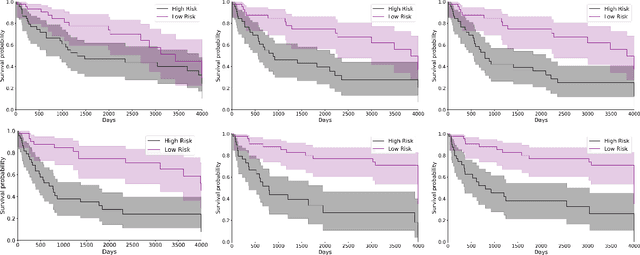
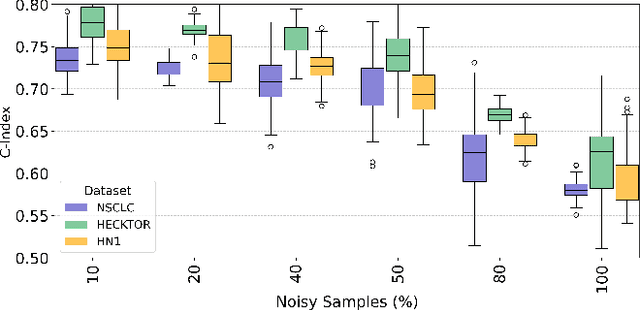
Abstract:Cancer survival prediction using multi-modal medical imaging presents a critical challenge in oncology, mainly due to the vulnerability of deep learning models to noise and protocol variations across imaging centers. Current approaches struggle to extract consistent features from heterogeneous CT and PET images, limiting their clinical applicability. We address these challenges by introducing RobSurv, a robust deep-learning framework that leverages vector quantization for resilient multi-modal feature learning. The key innovation of our approach lies in its dual-path architecture: one path maps continuous imaging features to learned discrete codebooks for noise-resistant representation, while the parallel path preserves fine-grained details through continuous feature processing. This dual representation is integrated through a novel patch-wise fusion mechanism that maintains local spatial relationships while capturing global context via Transformer-based processing. In extensive evaluations across three diverse datasets (HECKTOR, H\&N1, and NSCLC Radiogenomics), RobSurv demonstrates superior performance, achieving concordance index of 0.771, 0.742, and 0.734 respectively - significantly outperforming existing methods. Most notably, our model maintains robust performance even under severe noise conditions, with performance degradation of only 3.8-4.5\% compared to 8-12\% in baseline methods. These results, combined with strong generalization across different cancer types and imaging protocols, establish RobSurv as a promising solution for reliable clinical prognosis that can enhance treatment planning and patient care.
RibCageImp: A Deep Learning Framework for 3D Ribcage Implant Generation
Nov 14, 2024Abstract:The recovery of damaged or resected ribcage structures requires precise, custom-designed implants to restore the integrity and functionality of the thoracic cavity. Traditional implant design methods rely mainly on manual processes, making them time-consuming and susceptible to variability. In this work, we explore the feasibility of automated ribcage implant generation using deep learning. We present a framework based on 3D U-Net architecture that processes CT scans to generate patient-specific implant designs. To the best of our knowledge, this is the first investigation into automated thoracic implant generation using deep learning approaches. Our preliminary results, while moderate, highlight both the potential and the significant challenges in this complex domain. These findings establish a foundation for future research in automated ribcage reconstruction and identify key technical challenges that need to be addressed for practical implementation.
CoBooM: Codebook Guided Bootstrapping for Medical Image Representation Learning
Aug 08, 2024Abstract:Self-supervised learning (SSL) has emerged as a promising paradigm for medical image analysis by harnessing unannotated data. Despite their potential, the existing SSL approaches overlook the high anatomical similarity inherent in medical images. This makes it challenging for SSL methods to capture diverse semantic content in medical images consistently. This work introduces a novel and generalized solution that implicitly exploits anatomical similarities by integrating codebooks in SSL. The codebook serves as a concise and informative dictionary of visual patterns, which not only aids in capturing nuanced anatomical details but also facilitates the creation of robust and generalized feature representations. In this context, we propose CoBooM, a novel framework for self-supervised medical image learning by integrating continuous and discrete representations. The continuous component ensures the preservation of fine-grained details, while the discrete aspect facilitates coarse-grained feature extraction through the structured embedding space. To understand the effectiveness of CoBooM, we conduct a comprehensive evaluation of various medical datasets encompassing chest X-rays and fundus images. The experimental results reveal a significant performance gain in classification and segmentation tasks.
OPTiML: Dense Semantic Invariance Using Optimal Transport for Self-Supervised Medical Image Representation
Apr 18, 2024


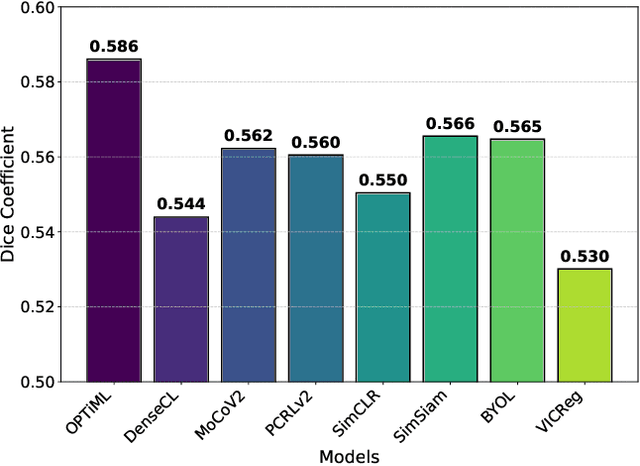
Abstract:Self-supervised learning (SSL) has emerged as a promising technique for medical image analysis due to its ability to learn without annotations. However, despite the promising potential, conventional SSL methods encounter limitations, including challenges in achieving semantic alignment and capturing subtle details. This leads to suboptimal representations, which fail to accurately capture the underlying anatomical structures and pathological details. In response to these constraints, we introduce a novel SSL framework OPTiML, employing optimal transport (OT), to capture the dense semantic invariance and fine-grained details, thereby enhancing the overall effectiveness of SSL in medical image representation learning. The core idea is to integrate OT with a cross-viewpoint semantics infusion module (CV-SIM), which effectively captures complex, fine-grained details inherent in medical images across different viewpoints. In addition to the CV-SIM module, OPTiML imposes the variance and covariance regularizations within OT framework to force the model focus on clinically relevant information while discarding less informative features. Through these, the proposed framework demonstrates its capacity to learn semantically rich representations that can be applied to various medical imaging tasks. To validate its effectiveness, we conduct experimental studies on three publicly available datasets from chest X-ray modality. Our empirical results reveal OPTiML's superiority over state-of-the-art methods across all evaluated tasks.
MLVICX: Multi-Level Variance-Covariance Exploration for Chest X-ray Self-Supervised Representation Learning
Mar 18, 2024Abstract:Self-supervised learning (SSL) is potentially useful in reducing the need for manual annotation and making deep learning models accessible for medical image analysis tasks. By leveraging the representations learned from unlabeled data, self-supervised models perform well on tasks that require little to no fine-tuning. However, for medical images, like chest X-rays, which are characterized by complex anatomical structures and diverse clinical conditions, there arises a need for representation learning techniques that can encode fine-grained details while preserving the broader contextual information. In this context, we introduce MLVICX (Multi-Level Variance-Covariance Exploration for Chest X-ray Self-Supervised Representation Learning), an approach to capture rich representations in the form of embeddings from chest X-ray images. Central to our approach is a novel multi-level variance and covariance exploration strategy that empowers the model to detect diagnostically meaningful patterns while reducing redundancy effectively. By enhancing the variance and covariance of the learned embeddings, MLVICX promotes the retention of critical medical insights by adapting both global and local contextual details. We demonstrate the performance of MLVICX in advancing self-supervised chest X-ray representation learning through comprehensive experiments. The performance enhancements we observe across various downstream tasks highlight the significance of the proposed approach in enhancing the utility of chest X-ray embeddings for precision medical diagnosis and comprehensive image analysis. For pertaining, we used the NIH-Chest X-ray dataset, while for downstream tasks, we utilized NIH-Chest X-ray, Vinbig-CXR, RSNA pneumonia, and SIIM-ACR Pneumothorax datasets. Overall, we observe more than 3% performance gains over SOTA SSL approaches in various downstream tasks.
Large Scale Time-Series Representation Learning via Simultaneous Low and High Frequency Feature Bootstrapping
Apr 24, 2022



Abstract:Learning representation from unlabeled time series data is a challenging problem. Most existing self-supervised and unsupervised approaches in the time-series domain do not capture low and high-frequency features at the same time. Further, some of these methods employ large scale models like transformers or rely on computationally expensive techniques such as contrastive learning. To tackle these problems, we propose a non-contrastive self-supervised learning approach efficiently captures low and high-frequency time-varying features in a cost-effective manner. Our method takes raw time series data as input and creates two different augmented views for two branches of the model, by randomly sampling the augmentations from same family. Following the terminology of BYOL, the two branches are called online and target network which allows bootstrapping of the latent representation. In contrast to BYOL, where a backbone encoder is followed by multilayer perceptron (MLP) heads, the proposed model contains additional temporal convolutional network (TCN) heads. As the augmented views are passed through large kernel convolution blocks of the encoder, the subsequent combination of MLP and TCN enables an effective representation of low as well as high-frequency time-varying features due to the varying receptive fields. The two modules (MLP and TCN) act in a complementary manner. We train an online network where each module learns to predict the outcome of the respective module of target network branch. To demonstrate the robustness of our model we performed extensive experiments and ablation studies on five real-world time-series datasets. Our method achieved state-of-art performance on all five real-world datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge