Advait Gosai
Audio MultiChallenge: A Multi-Turn Evaluation of Spoken Dialogue Systems on Natural Human Interaction
Dec 16, 2025



Abstract:End-to-end (E2E) spoken dialogue systems are increasingly replacing cascaded pipelines for voice-based human-AI interaction, processing raw audio directly without intermediate transcription. Existing benchmarks primarily evaluate these models on synthetic speech and single-turn tasks, leaving realistic multi-turn conversational ability underexplored. We introduce Audio MultiChallenge, an open-source benchmark to evaluate E2E spoken dialogue systems under natural multi-turn interaction patterns. Building on the text-based MultiChallenge framework, which evaluates Inference Memory, Instruction Retention, and Self Coherence, we introduce a new axis Voice Editing that tests robustness to mid-utterance speech repairs and backtracking. We further augment each axis to the audio modality, such as introducing Audio-Cue challenges for Inference Memory that require recalling ambient sounds and paralinguistic signals beyond semantic content. We curate 452 conversations from 47 speakers with 1,712 instance-specific rubrics through a hybrid audio-native agentic and human-in-the-loop pipeline that exposes model failures at scale while preserving natural disfluencies found in unscripted human speech. Our evaluation of proprietary and open-source models reveals that even frontier models struggle on our benchmark, with Gemini 3 Pro Preview (Thinking), our highest-performing model achieving a 54.65% pass rate. Error analysis shows that models fail most often on our new axes and that Self Coherence degrades with longer audio context. These failures reflect difficulty of tracking edits, audio cues, and long-range context in natural spoken dialogue. Audio MultiChallenge provides a reproducible testbed to quantify them and drive improvements in audio-native multi-turn interaction capability.
PRBench: Large-Scale Expert Rubrics for Evaluating High-Stakes Professional Reasoning
Nov 14, 2025Abstract:Frontier model progress is often measured by academic benchmarks, which offer a limited view of performance in real-world professional contexts. Existing evaluations often fail to assess open-ended, economically consequential tasks in high-stakes domains like Legal and Finance, where practical returns are paramount. To address this, we introduce Professional Reasoning Bench (PRBench), a realistic, open-ended, and difficult benchmark of real-world problems in Finance and Law. We open-source its 1,100 expert-authored tasks and 19,356 expert-curated criteria, making it, to our knowledge, the largest public, rubric-based benchmark for both legal and finance domains. We recruit 182 qualified professionals, holding JDs, CFAs, or 6+ years of experience, who contributed tasks inspired by their actual workflows. This process yields significant diversity, with tasks spanning 114 countries and 47 US jurisdictions. Our expert-curated rubrics are validated through a rigorous quality pipeline, including independent expert validation. Subsequent evaluation of 20 leading models reveals substantial room for improvement, with top scores of only 0.39 (Finance) and 0.37 (Legal) on our Hard subsets. We further catalog associated economic impacts of the prompts and analyze performance using human-annotated rubric categories. Our analysis shows that models with similar overall scores can diverge significantly on specific capabilities. Common failure modes include inaccurate judgments, a lack of process transparency and incomplete reasoning, highlighting critical gaps in their reliability for professional adoption.
Beyond Seeing: Evaluating Multimodal LLMs on Tool-Enabled Image Perception, Transformation, and Reasoning
Oct 14, 2025Abstract:Multimodal Large Language Models (MLLMs) are increasingly applied in real-world scenarios where user-provided images are often imperfect, requiring active image manipulations such as cropping, editing, or enhancement to uncover salient visual cues. Beyond static visual perception, MLLMs must also think with images: dynamically transforming visual content and integrating it with other tools to solve complex tasks. However, this shift from treating vision as passive context to a manipulable cognitive workspace remains underexplored. Most existing benchmarks still follow a think about images paradigm, where images are regarded as static inputs. To address this gap, we introduce IRIS, an Interactive Reasoning with Images and Systems that evaluates MLLMs' ability to perceive, transform, and reason across complex visual-textual tasks under the think with images paradigm. IRIS comprises 1,204 challenging, open-ended vision tasks (603 single-turn, 601 multi-turn) spanning across five diverse domains, each paired with detailed rubrics to enable systematic evaluation. Our evaluation shows that current MLLMs struggle with tasks requiring effective integration of vision and general-purpose tools. Even the strongest model (GPT-5-think) reaches only 18.68% pass rate. We further observe divergent tool-use behaviors, with OpenAI models benefiting from diverse image manipulations while Gemini-2.5-pro shows no improvement. By introducing the first benchmark centered on think with images, IRIS offers critical insights for advancing visual intelligence in MLLMs.
Automated Ensemble-Based Segmentation of Pediatric Brain Tumors: A Novel Approach Using the CBTN-CONNECT-ASNR-MICCAI BraTS-PEDs 2023 Challenge Data
Aug 14, 2023



Abstract:Brain tumors remain a critical global health challenge, necessitating advancements in diagnostic techniques and treatment methodologies. In response to the growing need for age-specific segmentation models, particularly for pediatric patients, this study explores the deployment of deep learning techniques using magnetic resonance imaging (MRI) modalities. By introducing a novel ensemble approach using ONet and modified versions of UNet, coupled with innovative loss functions, this study achieves a precise segmentation model for the BraTS-PEDs 2023 Challenge. Data augmentation, including both single and composite transformations, ensures model robustness and accuracy across different scanning protocols. The ensemble strategy, integrating the ONet and UNet models, shows greater effectiveness in capturing specific features and modeling diverse aspects of the MRI images which result in lesion_wise dice scores of 0.52, 0.72 and 0.78 for enhancing tumor, tumor core and whole tumor labels respectively. Visual comparisons further confirm the superiority of the ensemble method in accurate tumor region coverage. The results indicate that this advanced ensemble approach, building upon the unique strengths of individual models, offers promising prospects for enhanced diagnostic accuracy and effective treatment planning for brain tumors in pediatric brains.
Automated Ensemble-Based Segmentation of Adult Brain Tumors: A Novel Approach Using the BraTS AFRICA Challenge Data
Aug 14, 2023Abstract:Brain tumors, particularly glioblastoma, continue to challenge medical diagnostics and treatments globally. This paper explores the application of deep learning to multi-modality magnetic resonance imaging (MRI) data for enhanced brain tumor segmentation precision in the Sub-Saharan Africa patient population. We introduce an ensemble method that comprises eleven unique variations based on three core architectures: UNet3D, ONet3D, SphereNet3D and modified loss functions. The study emphasizes the need for both age- and population-based segmentation models, to fully account for the complexities in the brain. Our findings reveal that the ensemble approach, combining different architectures, outperforms single models, leading to improved evaluation metrics. Specifically, the results exhibit Dice scores of 0.82, 0.82, and 0.87 for enhancing tumor, tumor core, and whole tumor labels respectively. These results underline the potential of tailored deep learning techniques in precisely segmenting brain tumors and lay groundwork for future work to fine-tune models and assess performance across different brain regions.
Meta-Analysis of Transfer Learning for Segmentation of Brain Lesions
Jun 20, 2023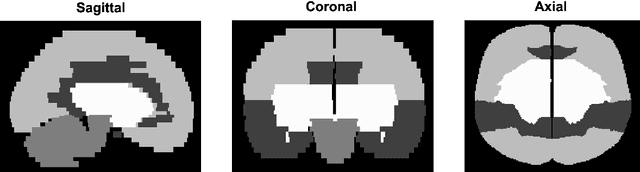
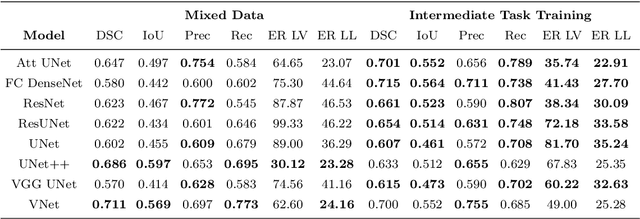
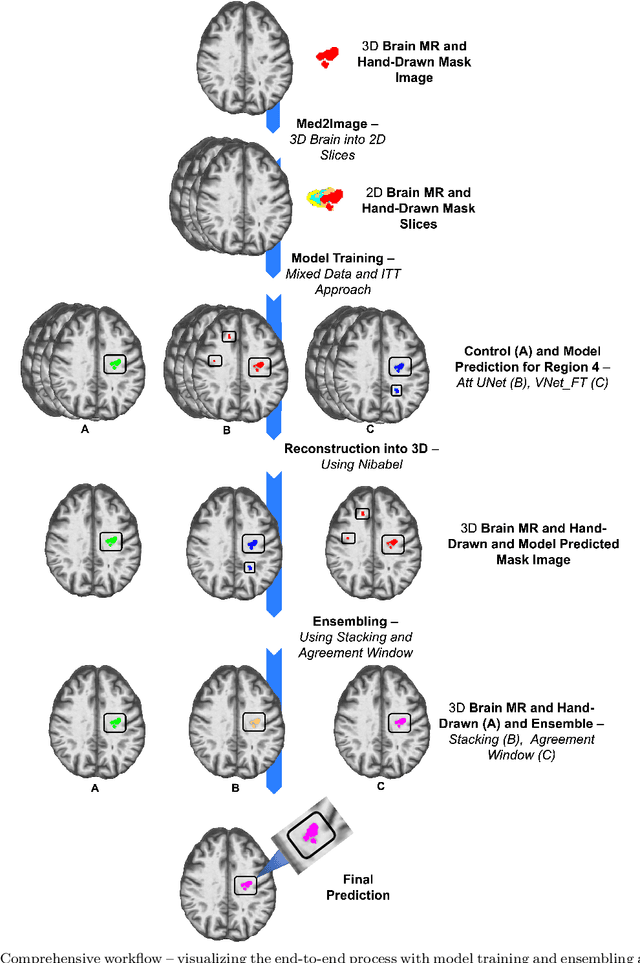
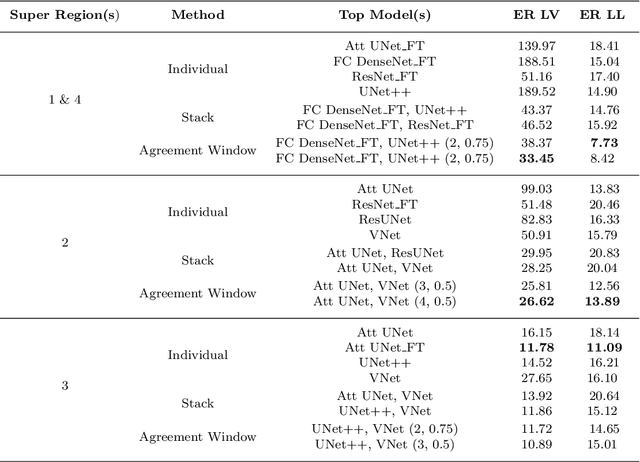
Abstract:A major challenge in stroke research and stroke recovery predictions is the determination of a stroke lesion's extent and its impact on relevant brain systems. Manual segmentation of stroke lesions from 3D magnetic resonance (MR) imaging volumes, the current gold standard, is not only very time-consuming, but its accuracy highly depends on the operator's experience. As a result, there is a need for a fully automated segmentation method that can efficiently and objectively measure lesion extent and the impact of each lesion to predict impairment and recovery potential which might be beneficial for clinical, translational, and research settings. We have implemented and tested a fully automatic method for stroke lesion segmentation which was developed using eight different 2D-model architectures trained via transfer learning (TL) and mixed data approaches. Additionally, the final prediction was made using a novel ensemble method involving stacking and agreement window. Our novel method was evaluated in a novel in-house dataset containing 22 T1w brain MR images, which were challenging in various perspectives, but mostly because they included T1w MR images from the subacute (which typically less well defined T1 lesions) and chronic stroke phase (which typically means well defined T1-lesions). Cross-validation results indicate that our new method can efficiently and automatically segment lesions fast and with high accuracy compared to ground truth. In addition to segmentation, we provide lesion volume and weighted lesion load of relevant brain systems based on the lesions' overlap with a canonical structural motor system that stretches from the cortical motor region to the lowest end of the brain stem.
SAM vs BET: A Comparative Study for Brain Extraction and Segmentation of Magnetic Resonance Images using Deep Learning
Apr 19, 2023Abstract:Brain extraction is a critical preprocessing step in various neuroimaging studies, particularly enabling accurate separation of brain from non-brain tissue and segmentation of relevant within-brain tissue compartments and structures using Magnetic Resonance Imaging (MRI) data. FSL's Brain Extraction Tool (BET), although considered the current gold standard for automatic brain extraction, presents limitations and can lead to errors such as over-extraction in brains with lesions affecting the outer parts of the brain, inaccurate differentiation between brain tissue and surrounding meninges, and susceptibility to image quality issues. Recent advances in computer vision research have led to the development of the Segment Anything Model (SAM) by Meta AI, which has demonstrated remarkable potential in zero-shot segmentation of objects in real-world scenarios. In the current paper, we present a comparative analysis of brain extraction techniques comparing SAM with a widely used and current gold standard technique called BET on a variety of brain scans with varying image qualities, MR sequences, and brain lesions affecting different brain regions. We find that SAM outperforms BET based on average Dice coefficient, IoU and accuracy metrics, particularly in cases where image quality is compromised by signal inhomogeneities, non-isotropic voxel resolutions, or the presence of brain lesions that are located near (or involve) the outer regions of the brain and the meninges. In addition, SAM has also unsurpassed segmentation properties allowing a fine grain separation of different issue compartments and different brain structures. These results suggest that SAM has the potential to emerge as a more accurate, robust and versatile tool for a broad range of brain extraction and segmentation applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge