Adrien Aumon
Forest-Guided Semantic Transport for Label-Supervised Manifold Alignment
Feb 01, 2026Abstract:Label-supervised manifold alignment bridges the gap between unsupervised and correspondence-based paradigms by leveraging shared label information to align multimodal datasets. Still, most existing methods rely on Euclidean geometry to model intra-domain relationships. This approach can fail when features are only weakly related to the task of interest, leading to noisy, semantically misleading structure and degraded alignment quality. To address this limitation, we introduce FoSTA (Forest-guided Semantic Transport Alignment), a scalable alignment framework that leverages forest-induced geometry to denoise intra-domain structure and recover task-relevant manifolds prior to alignment. FoSTA builds semantic representations directly from label-informed forest affinities and aligns them via fast, hierarchical semantic transport, capturing meaningful cross-domain relationships. Extensive comparisons with established baselines demonstrate that FoSTA improves correspondence recovery and label transfer on synthetic benchmarks and delivers strong performance in practical single-cell applications, including batch correction and biological conservation.
Scalable Tree Ensemble Proximities in Python
Jan 06, 2026Abstract:Tree ensemble methods such as Random Forests naturally induce supervised similarity measures through their decision tree structure, but existing implementations of proximities derived from tree ensembles typically suffer from quadratic time or memory complexity, limiting their scalability. In this work, we introduce a general framework for efficient proximity computation by defining a family of Separable Weighted Leaf-Collision Proximities. We show that any proximity measure in this family admits an exact sparse matrix factorization, restricting computation to leaf-level collisions and avoiding explicit pairwise comparisons. This formulation enables low-memory, scalable proximity computation using sparse linear algebra in Python. Empirical benchmarks demonstrate substantial runtime and memory improvements over traditional approaches, allowing tree ensemble proximities to scale efficiently to datasets with hundreds of thousands of samples on standard CPU hardware.
Random Forest Autoencoders for Guided Representation Learning
Feb 18, 2025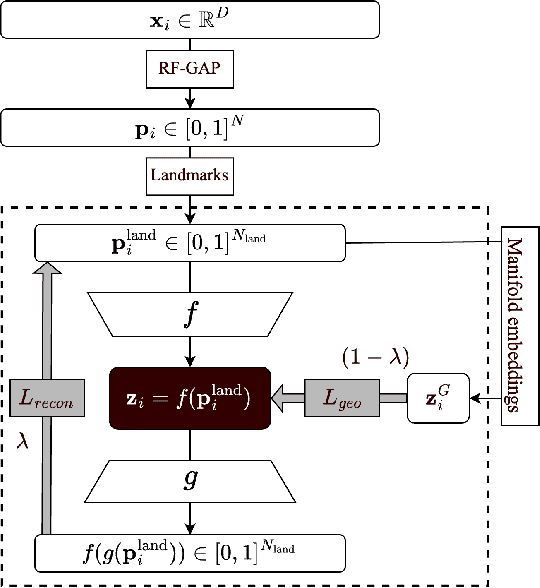
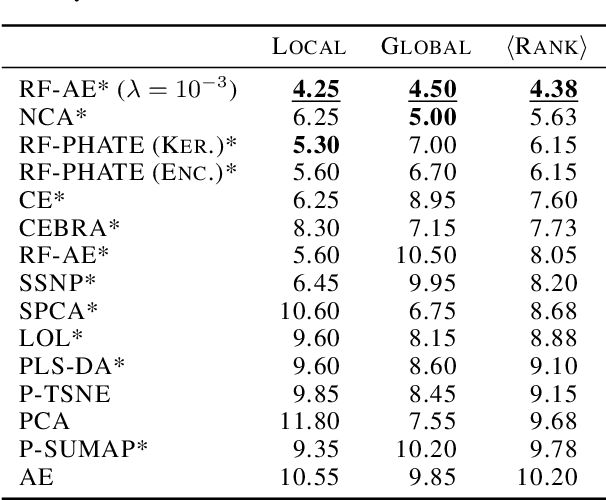
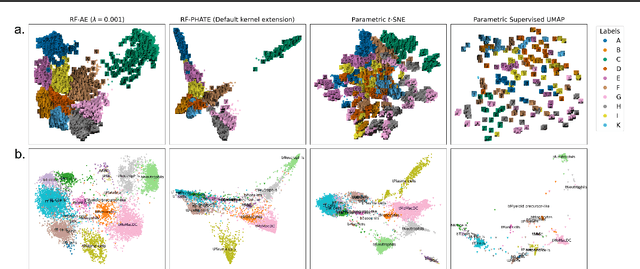
Abstract:Decades of research have produced robust methods for unsupervised data visualization, yet supervised visualization$\unicode{x2013}$where expert labels guide representations$\unicode{x2013}$remains underexplored, as most supervised approaches prioritize classification over visualization. Recently, RF-PHATE, a diffusion-based manifold learning method leveraging random forests and information geometry, marked significant progress in supervised visualization. However, its lack of an explicit mapping function limits scalability and prevents application to unseen data, posing challenges for large datasets and label-scarce scenarios. To overcome these limitations, we introduce Random Forest Autoencoders (RF-AE), a neural network-based framework for out-of-sample kernel extension that combines the flexibility of autoencoders with the supervised learning strengths of random forests and the geometry captured by RF-PHATE. RF-AE enables efficient out-of-sample supervised visualization and outperforms existing methods, including RF-PHATE's standard kernel extension, in both accuracy and interpretability. Additionally, RF-AE is robust to the choice of hyper-parameters and generalizes to any kernel-based dimensionality reduction method.
Enhancing Supervised Visualization through Autoencoder and Random Forest Proximities for Out-of-Sample Extension
Jun 06, 2024Abstract:The value of supervised dimensionality reduction lies in its ability to uncover meaningful connections between data features and labels. Common dimensionality reduction methods embed a set of fixed, latent points, but are not capable of generalizing to an unseen test set. In this paper, we provide an out-of-sample extension method for the random forest-based supervised dimensionality reduction method, RF-PHATE, combining information learned from the random forest model with the function-learning capabilities of autoencoders. Through quantitative assessment of various autoencoder architectures, we identify that networks that reconstruct random forest proximities are more robust for the embedding extension problem. Furthermore, by leveraging proximity-based prototypes, we achieve a 40% reduction in training time without compromising extension quality. Our method does not require label information for out-of-sample points, thus serving as a semi-supervised method, and can achieve consistent quality using only 10% of the training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge