Aayush Kumar Tyagi
MeasureNet: Measurement Based Celiac Disease Identification
Dec 02, 2024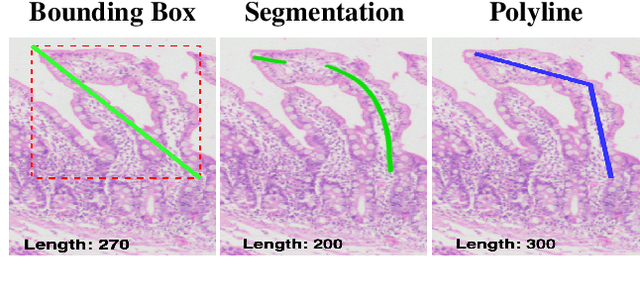
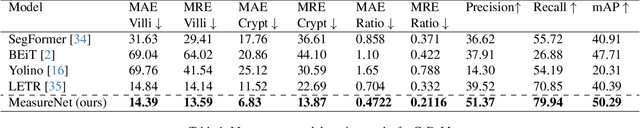
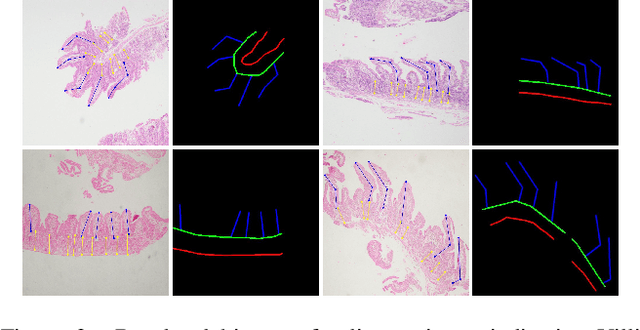
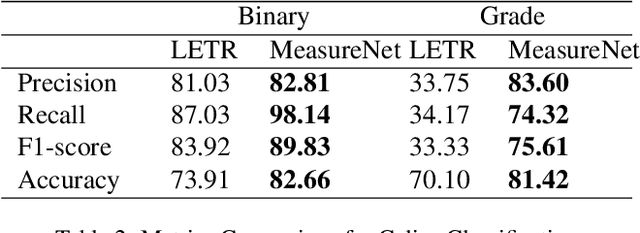
Abstract:Celiac disease is an autoimmune disorder triggered by the consumption of gluten. It causes damage to the villi, the finger-like projections in the small intestine that are responsible for nutrient absorption. Additionally, the crypts, which form the base of the villi, are also affected, impairing the regenerative process. The deterioration in villi length, computed as the villi-to-crypt length ratio, indicates the severity of celiac disease. However, manual measurement of villi-crypt length can be both time-consuming and susceptible to inter-observer variability, leading to inconsistencies in diagnosis. While some methods can perform measurement as a post-hoc process, they are prone to errors in the initial stages. This gap underscores the need for pathologically driven solutions that enhance measurement accuracy and reduce human error in celiac disease assessments. Our proposed method, MeasureNet, is a pathologically driven polyline detection framework incorporating polyline localization and object-driven losses specifically designed for measurement tasks. Furthermore, we leverage segmentation model to provide auxiliary guidance about crypt location when crypt are partially visible. To ensure that model is not overdependent on segmentation mask we enhance model robustness through a mask feature mixup technique. Additionally, we introduce a novel dataset for grading celiac disease, consisting of 750 annotated duodenum biopsy images. MeasureNet achieves an 82.66% classification accuracy for binary classification and 81% accuracy for multi-class grading of celiac disease. Code: https://github.com/dair-iitd/MeasureNet
Guided Prompting in SAM for Weakly Supervised Cell Segmentation in Histopathological Images
Nov 29, 2023Abstract:Cell segmentation in histopathological images plays a crucial role in understanding, diagnosing, and treating many diseases. However, data annotation for this is expensive since there can be a large number of cells per image, and expert pathologists are needed for labelling images. Instead, our paper focuses on using weak supervision -- annotation from related tasks -- to induce a segmenter. Recent foundation models, such as Segment Anything (SAM), can use prompts to leverage additional supervision during inference. SAM has performed remarkably well in natural image segmentation tasks; however, its applicability to cell segmentation has not been explored. In response, we investigate guiding the prompting procedure in SAM for weakly supervised cell segmentation when only bounding box supervision is available. We develop two workflows: (1) an object detector's output as a test-time prompt to SAM (D-SAM), and (2) SAM as pseudo mask generator over training data to train a standalone segmentation model (SAM-S). On finding that both workflows have some complementary strengths, we develop an integer programming-based approach to reconcile the two sets of segmentation masks, achieving yet higher performance. We experiment on three publicly available cell segmentation datasets namely, ConSep, MoNuSeg, and TNBC, and find that all SAM-based solutions hugely outperform existing weakly supervised image segmentation models, obtaining 9-15 pt Dice gains.
DeGPR: Deep Guided Posterior Regularization for Multi-Class Cell Detection and Counting
Apr 03, 2023Abstract:Multi-class cell detection and counting is an essential task for many pathological diagnoses. Manual counting is tedious and often leads to inter-observer variations among pathologists. While there exist multiple, general-purpose, deep learning-based object detection and counting methods, they may not readily transfer to detecting and counting cells in medical images, due to the limited data, presence of tiny overlapping objects, multiple cell types, severe class-imbalance, minute differences in size/shape of cells, etc. In response, we propose guided posterior regularization (DeGPR), which assists an object detector by guiding it to exploit discriminative features among cells. The features may be pathologist-provided or inferred directly from visual data. We validate our model on two publicly available datasets (CoNSeP and MoNuSAC), and on MuCeD, a novel dataset that we contribute. MuCeD consists of 55 biopsy images of the human duodenum for predicting celiac disease. We perform extensive experimentation with three object detection baselines on three datasets to show that DeGPR is model-agnostic, and consistently improves baselines obtaining up to 9% (absolute) mAP gains.
Unsupervised Domain Adaptation for Semantic Segmentation of NIR Images through Generative Latent Search
Jul 17, 2020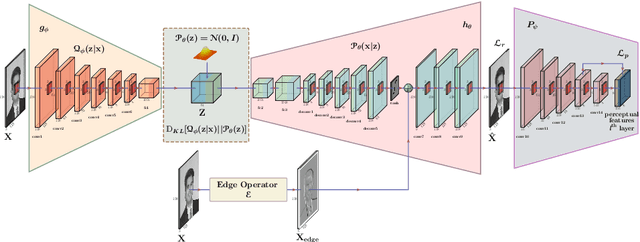


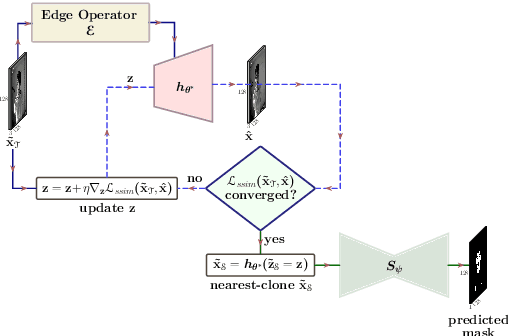
Abstract:Segmentation of the pixels corresponding to human skin is an essential first step in multiple applications ranging from surveillance to heart-rate estimation from remote-photoplethysmography. However, the existing literature considers the problem only in the visible-range of the EM-spectrum which limits their utility in low or no light settings where the criticality of the application is higher. To alleviate this problem, we consider the problem of skin segmentation from the Near-infrared images. However, Deep learning based state-of-the-art segmentation techniques demands large amounts of labelled data that is unavailable for the current problem. Therefore we cast the skin segmentation problem as that of target-independent Unsupervised Domain Adaptation (UDA) where we use the data from the Red-channel of the visible-range to develop skin segmentation algorithm on NIR images. We propose a method for target-independent segmentation where the 'nearest-clone' of a target image in the source domain is searched and used as a proxy in the segmentation network trained only on the source domain. We prove the existence of 'nearest-clone' and propose a method to find it through an optimization algorithm over the latent space of a Deep generative model based on variational inference. We demonstrate the efficacy of the proposed method for NIR skin segmentation over the state-of-the-art UDA segmentation methods on the two newly created skin segmentation datasets in NIR domain despite not having access to the target NIR data. Additionally, we report state-of-the-art results for adaption from Synthia to Cityscapes which is a popular setting in Unsupervised Domain Adaptation for semantic segmentation. The code and datasets are available at https://github.com/ambekarsameer96/GLSS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge